Temporal dynamics of stress response in Halomonas elongata to NaCl shock: physiological, metabolomic, and transcriptomic insights
- PMID: 38519954
- PMCID: PMC10960403
- DOI: 10.1186/s12934-024-02358-5
Temporal dynamics of stress response in Halomonas elongata to NaCl shock: physiological, metabolomic, and transcriptomic insights
Abstract
Background: The halophilic bacterium Halomonas elongata is an industrially important strain for ectoine production, with high value and intense research focus. While existing studies primarily delve into the adaptive mechanisms of this bacterium under fixed salt concentrations, there is a notable dearth of attention regarding its response to fluctuating saline environments. Consequently, the stress response of H. elongata to salt shock remains inadequately understood.
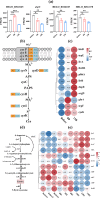
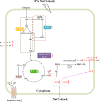
Results: This study investigated the stress response mechanism of H. elongata when exposed to NaCl shock at short- and long-time scales. Results showed that NaCl shock induced two major stresses, namely osmotic stress and oxidative stress. In response to the former, within the cell's tolerable range (1-8% NaCl shock), H. elongata urgently balanced the surging osmotic pressure by uptaking sodium and potassium ions and augmenting intracellular amino acid pools, particularly glutamate and glutamine. However, ectoine content started to increase until 20 min post-shock, rapidly becoming the dominant osmoprotectant, and reaching the maximum productivity (1450 ± 99 mg/L/h). Transcriptomic data also confirmed the delayed response in ectoine biosynthesis, and we speculate that this might be attributed to an intracellular energy crisis caused by NaCl shock. In response to oxidative stress, transcription factor cysB was significantly upregulated, positively regulating the sulfur metabolism and cysteine biosynthesis. Furthermore, the upregulation of the crucial peroxidase gene (HELO_RS18165) and the simultaneous enhancement of peroxidase (POD) and catalase (CAT) activities collectively constitute the antioxidant defense in H. elongata following shock. When exceeding the tolerance threshold of H. elongata (1-13% NaCl shock), the sustained compromised energy status, resulting from the pronounced inhibition of the respiratory chain and ATP synthase, may be a crucial factor leading to the stagnation of both cell growth and ectoine biosynthesis.
Conclusions: This study conducted a comprehensive analysis of H. elongata's stress response to NaCl shock at multiple scales. It extends the understanding of stress response of halophilic bacteria to NaCl shock and provides promising theoretical insights to guide future improvements in optimizing industrial ectoine production.
Keywords: Halomonas elongata; Ectoine; NaCl shock; Osmotic stress; Oxidative stress; Stress response.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

