Depletion of lamins B1 and B2 promotes chromatin mobility and induces differential gene expression by a mesoscale-motion-dependent mechanism
- PMID: 38519987
- PMCID: PMC10958841
- DOI: 10.1186/s13059-024-03212-y
Depletion of lamins B1 and B2 promotes chromatin mobility and induces differential gene expression by a mesoscale-motion-dependent mechanism
Abstract
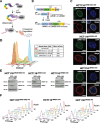
Background: B-type lamins are critical nuclear envelope proteins that interact with the three-dimensional genomic architecture. However, identifying the direct roles of B-lamins on dynamic genome organization has been challenging as their joint depletion severely impacts cell viability. To overcome this, we engineered mammalian cells to rapidly and completely degrade endogenous B-type lamins using Auxin-inducible degron technology.
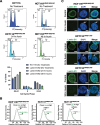
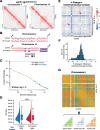
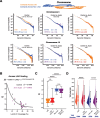
Results: Using live-cell Dual Partial Wave Spectroscopic (Dual-PWS) microscopy, Stochastic Optical Reconstruction Microscopy (STORM), in situ Hi-C, CRISPR-Sirius, and fluorescence in situ hybridization (FISH), we demonstrate that lamin B1 and lamin B2 are critical structural components of the nuclear periphery that create a repressive compartment for peripheral-associated genes. Lamin B1 and lamin B2 depletion minimally alters higher-order chromatin folding but disrupts cell morphology, significantly increases chromatin mobility, redistributes both constitutive and facultative heterochromatin, and induces differential gene expression both within and near lamin-associated domain (LAD) boundaries. Critically, we demonstrate that chromatin territories expand as upregulated genes within LADs radially shift inwards. Our results indicate that the mechanism of action of B-type lamins comes from their role in constraining chromatin motion and spatial positioning of gene-specific loci, heterochromatin, and chromatin domains.
Conclusions: Our findings suggest that, while B-type lamin degradation does not significantly change genome topology, it has major implications for three-dimensional chromatin conformation at the single-cell level both at the lamina-associated periphery and the non-LAD-associated nuclear interior with concomitant genome-wide transcriptional changes. This raises intriguing questions about the individual and overlapping roles of lamin B1 and lamin B2 in cellular function and disease.
Keywords: 3D chromatin organization; Auxin-inducible degron system; CRISPR-Sirius; in situ Hi-C; Lamin-associated domains; Nuclear lamina; Partial Wave Spectroscopic Microscopy; Topologically associated domains.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Update of
-
Depletion of lamins B1 and B2 alters chromatin mobility and induces differential gene expression by a mesoscale-motion dependent mechanism.bioRxiv [Preprint]. 2023 Jun 26:2023.06.26.546573. doi: 10.1101/2023.06.26.546573. bioRxiv. 2023. Update in: Genome Biol. 2024 Mar 22;25(1):77. doi: 10.1186/s13059-024-03212-y. PMID: 37425796 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- U54 CA261694/CA/NCI NIH HHS/United States
- UM1 HG009375/HG/NHGRI NIH HHS/United States
- U54 CA268084/NH/NIH HHS/United States
- T32AI083216/NH/NIH HHS/United States
- S10 OD026814/OD/NIH HHS/United States
- T32 AI083216/AI/NIAID NIH HHS/United States
- RM1 HG011016/HG/NHGRI NIH HHS/United States
- T32 GM142604/GM/NIGMS NIH HHS/United States
- S10 OD011996/OD/NIH HHS/United States
- R01 CA228272/CA/NCI NIH HHS/United States
- UM1 HG012649/HG/NHGRI NIH HHS/United States
- RM1HG011016-01A1/HG/NHGRI NIH HHS/United States
- R01CA228272/NH/NIH HHS/United States
- UM1HG009375/NH/NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- 5 UM1 HG012649/NH/NIH HHS/United States
- U54CA261694/NH/NIH HHS/United States
- P30 CA044579/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

