DNP-assisted solid-state NMR enables detection of proteins at nanomolar concentrations in fully protonated cellular milieu
- PMID: 38520488
- PMCID: PMC11572114
- DOI: 10.1007/s10858-024-00436-9
DNP-assisted solid-state NMR enables detection of proteins at nanomolar concentrations in fully protonated cellular milieu
Abstract
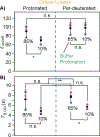
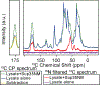
With the sensitivity enhancements conferred by dynamic nuclear polarization (DNP), magic angle spinning (MAS) solid state NMR spectroscopy experiments can attain the necessary sensitivity to detect very low concentrations of proteins. This potentially enables structural investigations of proteins at their endogenous levels in their biological contexts where their native stoichiometries with potential interactors is maintained. Yet, even with DNP, experiments are still sensitivity limited. Moreover, when an isotopically-enriched target protein is present at physiological levels, which typically range from low micromolar to nanomolar concentrations, the isotope content from the natural abundance isotopes in the cellular milieu can outnumber the isotope content of the target protein. Using isotopically enriched yeast prion protein, Sup35NM, diluted into natural abundance yeast lysates, we optimized sample composition. We found that modest cryoprotectant concentrations and fully protonated environments support efficient DNP. We experimentally validated theoretical calculations of the limit of specificity for an isotopically enriched protein in natural abundance cellular milieu. We establish that, using pulse sequences that are selective for adjacent NMR-active nuclei, proteins can be specifically detected in cellular milieu at concentrations in the hundreds of nanomolar. Finally, we find that maintaining native stoichiometries of the protein of interest to the components of the cellular environment may be important for proteins that make specific interactions with cellular constituents.
Keywords: DNP solid-state NMR; In-cell NMR; Sup35; Yeast prions.
© 2024. The Author(s), under exclusive licence to Springer Nature B.V.
Figures



Similar articles
-
Sensitivity-enhanced NMR reveals alterations in protein structure by cellular milieus.Cell. 2015 Oct 22;163(3):620-8. doi: 10.1016/j.cell.2015.09.024. Epub 2015 Oct 8. Cell. 2015. PMID: 26456111 Free PMC article.
-
Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):3642-3647. doi: 10.1073/pnas.1619051114. Epub 2017 Mar 22. Proc Natl Acad Sci U S A. 2017. PMID: 28330994 Free PMC article.
-
DNP-Assisted NMR Investigation of Proteins at Endogenous Levels in Cellular Milieu.Methods Enzymol. 2019;615:373-406. doi: 10.1016/bs.mie.2018.08.023. Epub 2018 Sep 18. Methods Enzymol. 2019. PMID: 30638534 Free PMC article.
-
Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review.
-
New NMR tools for protein structure and function: Spin tags for dynamic nuclear polarization solid state NMR.Arch Biochem Biophys. 2017 Aug 15;628:102-113. doi: 10.1016/j.abb.2017.06.010. Epub 2017 Jun 13. Arch Biochem Biophys. 2017. PMID: 28623034 Free PMC article. Review.
Cited by
-
Probing Biomolecular Interactions with Paramagnetic Nuclear Magnetic Resonance Spectroscopy.Chembiochem. 2025 Mar 15;26(6):e202400903. doi: 10.1002/cbic.202400903. Epub 2025 Jan 13. Chembiochem. 2025. PMID: 39803829 Free PMC article. Review.
-
Spatially resolved DNP-assisted NMR illuminates the conformational ensemble of α-synuclein in intact viable cells.bioRxiv [Preprint]. 2025 Jan 4:2023.10.24.563877. doi: 10.1101/2023.10.24.563877. bioRxiv. 2025. Update in: Proc Natl Acad Sci U S A. 2025 Jun 10;122(23):e2500367122. doi: 10.1073/pnas.2500367122. PMID: 37961511 Free PMC article. Updated. Preprint.
-
Sorbitol-Based Glass Matrices Enable Dynamic Nuclear Polarization beyond 200 K.J Phys Chem Lett. 2024 Aug 29;15(34):8743-8751. doi: 10.1021/acs.jpclett.4c02054. Epub 2024 Aug 20. J Phys Chem Lett. 2024. PMID: 39162721 Free PMC article.
-
Stability of the polarization agent AsymPolPOK in intact and lysed mammalian cells.bioRxiv [Preprint]. 2024 Nov 11:2024.11.09.622814. doi: 10.1101/2024.11.09.622814. bioRxiv. 2024. Update in: J Magn Reson. 2025 May;374:107864. doi: 10.1016/j.jmr.2025.107864. PMID: 39605460 Free PMC article. Updated. Preprint.
-
In cell NMR reveals cells selectively amplify and structurally remodel amyloid fibrils.bioRxiv [Preprint]. 2024 Sep 10:2024.09.09.612142. doi: 10.1101/2024.09.09.612142. bioRxiv. 2024. PMID: 39314304 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

