A genetic screen identifies a role for oprF in Pseudomonas aeruginosa biofilm stimulation by subinhibitory antibiotics
- PMID: 38521769
- PMCID: PMC10960818
- DOI: 10.1038/s41522-024-00496-7
A genetic screen identifies a role for oprF in Pseudomonas aeruginosa biofilm stimulation by subinhibitory antibiotics
Abstract
Biofilms are surface-associated communities of bacteria that grow in a self-produced matrix of polysaccharides, proteins, and extracellular DNA (eDNA). Sub-minimal inhibitory concentrations (sub-MIC) of antibiotics induce biofilm formation, potentially as a defensive response to antibiotic stress. However, the mechanisms behind sub-MIC antibiotic-induced biofilm formation are unclear. We show that treatment of Pseudomonas aeruginosa with multiple classes of sub-MIC antibiotics with distinct targets induces biofilm formation. Further, addition of exogenous eDNA or cell lysate failed to increase biofilm formation to the same extent as antibiotics, suggesting that the release of cellular contents by antibiotic-driven bacteriolysis is insufficient. Using a genetic screen for stimulation-deficient mutants, we identified the outer membrane porin OprF and the ECF sigma factor SigX as important. Similarly, loss of OmpA - the Escherichia coli OprF homolog - prevented sub-MIC antibiotic stimulation of E. coli biofilms. Our screen also identified the periplasmic disulfide bond-forming enzyme DsbA and a predicted cyclic-di-GMP phosphodiesterase encoded by PA2200 as essential for biofilm stimulation. The phosphodiesterase activity of PA2200 is likely controlled by a disulfide bond in its regulatory domain, and folding of OprF is influenced by disulfide bond formation, connecting the mutant phenotypes. Addition of reducing agent dithiothreitol prevented sub-MIC antibiotic biofilm stimulation. Finally, activation of a c-di-GMP-responsive promoter follows treatment with sub-MIC antibiotics in the wild-type but not an oprF mutant. Together, these results show that antibiotic-induced biofilm formation is likely driven by a signaling pathway that translates changes in periplasmic redox state into elevated biofilm formation through increases in c-di-GMP.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
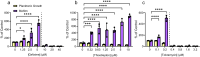
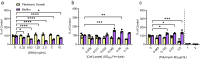

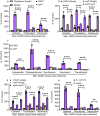

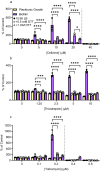
References
-
- Zilberberg MD, Shorr AF. Prevalence of multidrug-resistant Pseudomonas aeruginosa and carbapenem-resistant Enterobacteriaceae among specimens from hospitalized patients with pneumonia and bloodstream infections in the United States from 2000 to 2009. J. Hosp. Med. 2013;8:559–563. doi: 10.1002/jhm.2080. - DOI - PubMed
-
- Tacconelli E, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2017;17:1–10. - PubMed
MeSH terms
Substances
Grants and funding
- RGPIN-2021-04237/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- PGS-D/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- USRA/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
LinkOut - more resources
Full Text Sources
Medical

