Developing a machine learning model for accurate nucleoside hydrogels prediction based on descriptors
- PMID: 38521777
- PMCID: PMC10960799
- DOI: 10.1038/s41467-024-46866-9
Developing a machine learning model for accurate nucleoside hydrogels prediction based on descriptors
Abstract
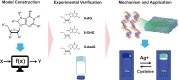
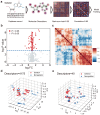
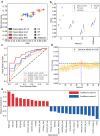
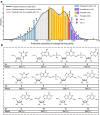
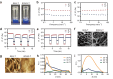
Supramolecular hydrogels derived from nucleosides have been gaining significant attention in the biomedical field due to their unique properties and excellent biocompatibility. However, a major challenge in this field is that there is no model for predicting whether nucleoside derivative will form a hydrogel. Here, we successfully develop a machine learning model to predict the hydrogel-forming ability of nucleoside derivatives. The optimal model with a 71% (95% Confidence Interval, 0.69-0.73) accuracy is established based on a dataset of 71 reported nucleoside derivatives. 24 molecules are selected via the optimal model external application and the hydrogel-forming ability is experimentally verified. Among these, two rarely reported cation-independent nucleoside hydrogels are found. Based on their self-assemble mechanisms, the cation-independent hydrogel is found to have potential applications in rapid visual detection of Ag+ and cysteine. Here, we show the machine learning model may provide a tool to predict nucleoside derivatives with hydrogel-forming ability.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Nucleoside-Based Supramolecular Hydrogels: From Synthesis and Structural Properties to Biomedical and Tissue Engineering Applications.ACS Biomater Sci Eng. 2023 Jan 9;9(1):40-61. doi: 10.1021/acsbiomaterials.2c01051. Epub 2022 Dec 16. ACS Biomater Sci Eng. 2023. PMID: 36524860 Review.
-
Multiresponsive Supramolecular Luminescent Hydrogels Based on a Nucleoside/Lanthanide Complex.ACS Appl Mater Interfaces. 2019 Dec 18;11(50):47404-47412. doi: 10.1021/acsami.9b17236. Epub 2019 Dec 6. ACS Appl Mater Interfaces. 2019. PMID: 31763814
-
High-Strength and Injectable Supramolecular Hydrogel Self-Assembled by Monomeric Nucleoside for Tooth-Extraction Wound Healing.Adv Mater. 2022 Apr;34(13):e2108300. doi: 10.1002/adma.202108300. Epub 2022 Feb 14. Adv Mater. 2022. PMID: 35066934
-
Self-Assembly of Metallo-Nucleoside Hydrogels for Injectable Materials That Promote Wound Closure.ACS Appl Mater Interfaces. 2019 Jun 5;11(22):19743-19750. doi: 10.1021/acsami.9b02265. Epub 2019 May 22. ACS Appl Mater Interfaces. 2019. PMID: 31081327
-
Supramolecular gels made from nucleobase, nucleoside and nucleotide analogs.Chem Soc Rev. 2016 Jun 7;45(11):3188-206. doi: 10.1039/c6cs00183a. Epub 2016 May 5. Chem Soc Rev. 2016. PMID: 27146863 Review.
Cited by
-
AI-driven 3D bioprinting for regenerative medicine: From bench to bedside.Bioact Mater. 2024 Nov 23;45:201-230. doi: 10.1016/j.bioactmat.2024.11.021. eCollection 2025 Mar. Bioact Mater. 2024. PMID: 39651398 Free PMC article. Review.
-
Integration of deep neural network modeling and LC-MS-based pseudo-targeted metabolomics to discriminate easily confused ginseng species.J Pharm Anal. 2025 Jan;15(1):101116. doi: 10.1016/j.jpha.2024.101116. Epub 2024 Sep 26. J Pharm Anal. 2025. PMID: 39902459 Free PMC article.
-
Integrating machine learning for the optimization of polyacrylamide/alginate hydrogel.Regen Biomater. 2024 Sep 2;11:rbae109. doi: 10.1093/rb/rbae109. eCollection 2024. Regen Biomater. 2024. PMID: 39323746 Free PMC article.
-
Clustering and classification for dry bean feature imbalanced data.Sci Rep. 2024 Dec 28;14(1):31058. doi: 10.1038/s41598-024-82253-6. Sci Rep. 2024. PMID: 39730714 Free PMC article.
-
Enhancing fluorescent probe design through multilayer interaction convolutional networks: advancing biosensing and bioimaging precision.Chem Sci. 2025 Apr 22;16(20):8853-8860. doi: 10.1039/d4sc08695c. eCollection 2025 May 21. Chem Sci. 2025. PMID: 40271040 Free PMC article.
References
-
- Bang I. Examination or the guanyle acid. Biochem. Z. 1910;26:293–311.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

