Expression of USP25 associates with fibrosis, inflammation and metabolism changes in IgG4-related disease
- PMID: 38521787
- PMCID: PMC10960850
- DOI: 10.1038/s41467-024-45977-7
Expression of USP25 associates with fibrosis, inflammation and metabolism changes in IgG4-related disease
Abstract
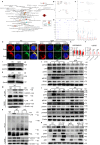
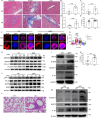
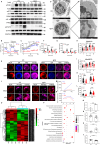
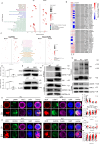
IgG4-related disease (IgG4-RD) has complex clinical manifestations ranging from fibrosis and inflammation to deregulated metabolism. The molecular mechanisms underpinning these phenotypes are unclear. In this study, by using IgG4-RD patient peripheral blood mononuclear cells (PBMCs), IgG4-RD cell lines and Usp25 knockout mice, we show that ubiquitin-specific protease 25 (USP25) engages in multiple pathways to regulate fibrotic and inflammatory pathways that are characteristic to IgG4-RD. Reduced USP25 expression in IgG4-RD leads to increased SMAD3 activation, which contributes to fibrosis and induces inflammation through the IL-1β inflammatory axis. Mechanistically, USP25 prevents ubiquitination of RAC1, thus, downregulation of USP25 leads to ubiquitination and degradation of RAC1. Decreased RAC1 levels result in reduced aldolase A release from the actin cytoskeleton, which then lowers glycolysis. The expression of LYN, a component of the B cell receptor signalosome is also reduced in USP25-deficient B cells, which might result in B cell activation deficiency. Altogether, our results indicate a potential anti-inflammatory and anti-fibrotic role for USP25 and make USP25 a promising diagnostic marker and potential therapeutic target in IgG4-RD.
© 2024. The Author(s).
Conflict of interest statement
Xin.Zh., Xia.Zh., and J.L. were employed by GeneMind Biosciences Company Limited, Shenzhen, China. Heather Miller was employed by Cytek Biosciences, R&D Clinical Reagents, Fremont, CA, United States. The remaining authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

