An evaluation of the replicability of analyses using synthetic health data
- PMID: 38521806
- PMCID: PMC10960851
- DOI: 10.1038/s41598-024-57207-7
An evaluation of the replicability of analyses using synthetic health data
Abstract
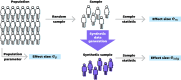
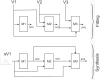
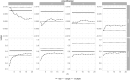
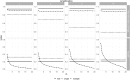
Synthetic data generation is being increasingly used as a privacy preserving approach for sharing health data. In addition to protecting privacy, it is important to ensure that generated data has high utility. A common way to assess utility is the ability of synthetic data to replicate results from the real data. Replicability has been defined using two criteria: (a) replicate the results of the analyses on real data, and (b) ensure valid population inferences from the synthetic data. A simulation study using three heterogeneous real-world datasets evaluated the replicability of logistic regression workloads. Eight replicability metrics were evaluated: decision agreement, estimate agreement, standardized difference, confidence interval overlap, bias, confidence interval coverage, statistical power, and precision (empirical SE). The analysis of synthetic data used a multiple imputation approach whereby up to 20 datasets were generated and the fitted logistic regression models were combined using combining rules for fully synthetic datasets. The effects of synthetic data amplification were evaluated, and two types of generative models were used: sequential synthesis using boosted decision trees and a generative adversarial network (GAN). Privacy risk was evaluated using a membership disclosure metric. For sequential synthesis, adjusted model parameters after combining at least ten synthetic datasets gave high decision and estimate agreement, low standardized difference, as well as high confidence interval overlap, low bias, the confidence interval had nominal coverage, and power close to the nominal level. Amplification had only a marginal benefit. Confidence interval coverage from a single synthetic dataset without applying combining rules were erroneous, and statistical power, as expected, was artificially inflated when amplification was used. Sequential synthesis performed considerably better than the GAN across multiple datasets. Membership disclosure risk was low for all datasets and models. For replicable results, the statistical analysis of fully synthetic data should be based on at least ten generated datasets of the same size as the original whose analyses results are combined. Analysis results from synthetic data without applying combining rules can be misleading. Replicability results are dependent on the type of generative model used, with our study suggesting that sequential synthesis has good replicability characteristics for common health research workloads.
© 2024. The Author(s).
Conflict of interest statement
This work was performed in collaboration with Replica Analytics Ltd. This company is a spin-off from the Children’s Hospital of Eastern Ontario Research Institute. KEE is co-founder and has equity in this company. LM and XF are data scientists employed by Replica Analytics Ltd. AAH has no competing interests.
Figures


References
-
- Wang, Z., Myles, P. & Tucker, A. Generating and evaluating synthetic UK primary care data: Preserving data utility patient privacy. In 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Cordoba. 126–31. 10.1109/CBMS.2019.00036 (2019).
-
- Wang Z, Myles P, Tucker A. Generating and evaluating cross-sectional synthetic electronic healthcare data: Preserving data utility and patient privacy. Comput. Intell. 2021;37:819–851. doi: 10.1111/coin.12427. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

