Homozygous missense variants in YKT6 result in loss of function and are associated with developmental delay, with or without severe infantile liver disease and risk for hepatocellular carcinoma
- PMID: 38522068
- PMCID: PMC11335040
- DOI: 10.1016/j.gim.2024.101125
Homozygous missense variants in YKT6 result in loss of function and are associated with developmental delay, with or without severe infantile liver disease and risk for hepatocellular carcinoma
Abstract
Purpose: YKT6 plays important roles in multiple intracellular vesicle trafficking events but has not been associated with Mendelian diseases.
Methods: We report 3 unrelated individuals with rare homozygous missense variants in YKT6 who exhibited neurological disease with or without a progressive infantile liver disease. We modeled the variants in Drosophila. We generated wild-type and variant genomic rescue constructs of the fly ortholog dYkt6 and compared their ability in rescuing the loss-of-function phenotypes in mutant flies. We also generated a dYkt6KozakGAL4 allele to assess the expression pattern of dYkt6.
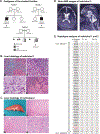
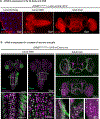
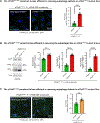
Results: Two individuals are homozygous for YKT6 [NM_006555.3:c.554A>G p.(Tyr185Cys)] and exhibited normal prenatal course followed by failure to thrive, developmental delay, and progressive liver disease. Haplotype analysis identified a shared homozygous region flanking the variant, suggesting a common ancestry. The third individual is homozygous for YKT6 [NM_006555.3:c.191A>G p.(Tyr64Cys)] and exhibited neurodevelopmental disorders and optic atrophy. Fly dYkt6 is essential and is expressed in the fat body (analogous to liver) and central nervous system. Wild-type genomic rescue constructs can rescue the lethality and autophagic flux defects, whereas the variants are less efficient in rescuing the phenotypes.
Conclusion: The YKT6 variants are partial loss-of-function alleles, and the p.(Tyr185Cys) is more severe than p.(Tyr64Cys).
Keywords: Autophagy; Drosophila; Failure to thrive; Fat body; Syrian Christians of India.
Copyright © 2024 American College of Medical Genetics and Genomics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest BCM and Miraca Holdings have formed a joint venture with shared ownership and governance of BG, which performs clinical microarray analysis, clinical ES (cES), and clinical biochemical studies. James R. Lupski serves on the Scientific Advisory Board of the BG. James R. Lupski has stock ownership in 23andMe, is a paid consultant for Genomics International, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, genomic disorders, and bacterial genomic fingerprinting. Nhu Thao Nguyen Galván serves as a consultant for 3DSystems. Davut Pehlivan provides consulting service for Ionis Pharmaceuticals. Wendy K. Chung is on the Board of Directors of Prime Medicine and Rallybio. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories. All other authors declare no conflicts of interest.
Figures


Similar articles
-
Functional interpretation of ATAD3A variants in neuro-mitochondrial phenotypes.Genome Med. 2021 Apr 12;13(1):55. doi: 10.1186/s13073-021-00873-3. Genome Med. 2021. PMID: 33845882 Free PMC article.
-
Deficiencies in vesicular transport mediated by TRAPPC4 are associated with severe syndromic intellectual disability.Brain. 2020 Jan 1;143(1):112-130. doi: 10.1093/brain/awz374. Brain. 2020. PMID: 31794024 Free PMC article.
-
TNPO2 variants associate with human developmental delays, neurologic deficits, and dysmorphic features and alter TNPO2 activity in Drosophila.Am J Hum Genet. 2021 Sep 2;108(9):1669-1691. doi: 10.1016/j.ajhg.2021.06.019. Epub 2021 Jul 26. Am J Hum Genet. 2021. PMID: 34314705 Free PMC article.
-
A novel pathogenic variant in the 3' end of the AGTPBP1 gene gives rise to neurodegeneration without cerebellar atrophy: an expansion of the disease phenotype?Neurogenetics. 2021 May;22(2):127-132. doi: 10.1007/s10048-021-00643-8. Epub 2021 Apr 28. Neurogenetics. 2021. PMID: 33909173 Review.
-
Homozygous variants in pyrroline-5-carboxylate reductase 2 (PYCR2) in patients with progressive microcephaly and hypomyelinating leukodystrophy.Am J Med Genet A. 2017 Feb;173(2):460-470. doi: 10.1002/ajmg.a.38049. Epub 2016 Nov 11. Am J Med Genet A. 2017. PMID: 27860360 Review.
References
Web resources
-
- AlphaFold: https://alphafold.ebi.ac.uk/
-
- MARRVEL: http://marrvel.org/
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

