Design and validation of a reporter mouse to study the dynamic regulation of TFEB and TFE3 activity through in vivo imaging techniques
- PMID: 38522425
- PMCID: PMC11262230
- DOI: 10.1080/15548627.2024.2334111
Design and validation of a reporter mouse to study the dynamic regulation of TFEB and TFE3 activity through in vivo imaging techniques
Abstract
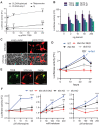
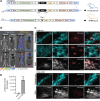
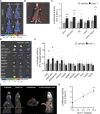
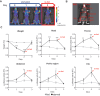
TFEB and TFE3 belong to the MiT/TFE family of transcription factors that bind identical DNA responsive elements in the regulatory regions of target genes. They are involved in regulating lysosomal biogenesis, function, exocytosis, autophagy, and lipid catabolism. Precise control of TFEB and TFE3 activity is crucial for processes such as senescence, stress response, energy metabolism, and cellular catabolism. Dysregulation of these factors is implicated in various diseases, thus researchers have explored pharmacological approaches to modulate MiT/TFE activity, considering these transcription factors as potential therapeutic targets. However, the physiological complexity of their functions and the lack of suitable in vivo tools have limited the development of selective MiT/TFE modulating agents. Here, we have created a reporter-based biosensor, named CLEARoptimized, facilitating the pharmacological profiling of TFEB- and TFE3-mediated transcription. This innovative tool enables the measurement of TFEB and TFE3 activity in living cells and mice through imaging and biochemical techniques. CLEARoptimized consists of a promoter with six coordinated lysosomal expression and regulation motifs identified through an in-depth bioinformatic analysis of the promoters of 128 TFEB-target genes. The biosensor drives the expression of luciferase and tdTomato reporter genes, allowing the quantification of TFEB and TFE3 activity in cells and in animals through optical imaging and biochemical assays. The biosensor's validity was confirmed by modulating MiT/TFE activity in both cell culture and reporter mice using physiological and pharmacological stimuli. Overall, this study introduces an innovative tool for studying autophagy and lysosomal pathway modulation at various biological levels, from individual cells to the entire organism.Abbreviations: CLEAR: coordinated lysosomal expression and regulation; MAR: matrix attachment regions; MiT: microphthalmia-associated transcription factor; ROI: region of interest; TBS: tris-buffered saline; TF: transcription factor; TFE3: transcription factor binding to IGHM enhancer 3; TFEB: transcription factor EB; TH: tyrosine hydroxylase; TK: thymidine kinase; TSS: transcription start site.
Keywords: Autophagy; drug discovery; luciferase; lysosomal pathway; non-invasive study of TFEB and TFE3; optical imaging.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
