Network switches and their role in circadian clocks
- PMID: 38522517
- PMCID: PMC11044057
- DOI: 10.1016/j.jbc.2024.107220
Network switches and their role in circadian clocks
Abstract
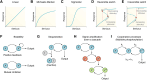
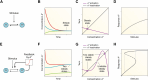
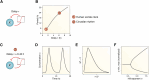
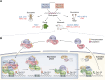
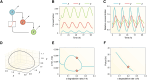
Circadian rhythms are generated by complex interactions among genes and proteins. Self-sustained ∼24 h oscillations require negative feedback loops and sufficiently strong nonlinearities that are the product of molecular and network switches. Here, we review common mechanisms to obtain switch-like behavior, including cooperativity, antagonistic enzymes, multisite phosphorylation, positive feedback, and sequestration. We discuss how network switches play a crucial role as essential components in cellular circadian clocks, serving as integral parts of transcription-translation feedback loops that form the basis of circadian rhythm generation. The design principles of network switches and circadian clocks are illustrated by representative mathematical models that include bistable systems and negative feedback loops combined with Hill functions. This work underscores the importance of negative feedback loops and network switches as essential design principles for biological oscillations, emphasizing how an understanding of theoretical concepts can provide insights into the mechanisms generating biological rhythms.
Keywords: bistability; circadian clock; feedback loops; mathematical models; network switches; ultrasensitivity.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Okamura H., Aramburu J., Garcıa-Rodrıguez C., Viola J.P.B., Raghavan A., Tahiliani M., et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell. 2000;6:539–550. - PubMed
-
- Salazar C., Höfer T. Allosteric regulation of the transcription factor NFAT1 by multiple phosphorylation sites: a mathematical analysis. J. Mol. Biol. 2003;327:31–45. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

