Detergent-induced quantitatively limited formation of diacyl phosphatidylinositol dimannoside in Mycobacterium smegmatis
- PMID: 38522749
- PMCID: PMC11269278
- DOI: 10.1016/j.jlr.2024.100533
Detergent-induced quantitatively limited formation of diacyl phosphatidylinositol dimannoside in Mycobacterium smegmatis
Abstract
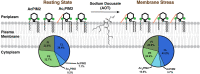
Mycobacterial plasma membrane, together with the peptidoglycan-arabinogalactan cell wall and waxy outer membrane, creates a robust permeability barrier against xenobiotics. The fact that several antituberculosis drugs target plasma membrane-embedded enzymes underscores the importance of the plasma membrane in bacterial physiology and pathogenesis. Nevertheless, its accurate phospholipid composition remains undefined, with conflicting reports on the abundance of phosphatidylinositol mannosides (PIMs), physiologically important glycolipids evolutionarily conserved among mycobacteria and related bacteria. Some studies indicate cardiolipin, phosphatidylethanolamine, and phosphatidylinositol as dominant structural phospholipids. Conversely, some suggest PIMs dominate the plasma membrane. A striking example of the latter is the use of reverse micelle extraction, showing diacyl phosphatidylinositol dimannoside (Ac2PIM2) as the most abundant phospholipid in a model organism, Mycobacterium smegmatis. Our recent work reveals a rapid response mechanism to membrane-fluidizing stress in mycobacterial plasma membrane: monoacyl phosphatidylinositol dimannoside and hexamannoside (AcPIM2 and AcPIM6) are converted to diacyl forms (Ac2PIM2 and Ac2PIM6). Given the dynamic nature of PIMs, we aimed to resolve the conflicting data in the literature. We show that unstressed M. smegmatis lacks an Ac2PIM2-dominated plasma membrane. Ac2PIM2 accumulation is induced by experimental conditions involving sodium docusate, a component of the reverse micellar solution. Using chemically synthesized PIMs as standards, we accurately quantified phospholipid ratio in M. smegmatis through liquid chromatography-mass spectrometry, revealing that mycobacterial plasma membrane is dominated by cardiolipin, phosphatidylethanolamine, and phosphatidylinositol. PIMs are quantitatively minor but responsive to environmental stresses in M. smegmatis. Our study paves the way for accurate modeling of mycobacterial plasma membrane.
Keywords: biosynthesis; chemical synthesis; glycolipids; membrane fluidity; metabolism; phospholipids; reverse micelle extraction; stress response.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures







References
-
- LaBelle Y.S., Walker R.W. The phospholipids of Mycobacterium smegmatis. Am. Rev. Respir. Dis. 1972;105:625–628. - PubMed
-
- Jackson M., Crick D.C., Brennan P.J. Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 2000;275:30092–30099. - PubMed
-
- Akamatsu Y., Ono Y., Nojima S. Phospholipid patterns in subcellular fractions of Mycobacterium phlei. J. Biochem. 1966;59:176–182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

