Single-Molecule Characterization and Super-Resolution Imaging of Alzheimer's Disease-Relevant Tau Aggregates in Human Samples
- PMID: 38523073
- PMCID: PMC11497306
- DOI: 10.1002/anie.202317756
Single-Molecule Characterization and Super-Resolution Imaging of Alzheimer's Disease-Relevant Tau Aggregates in Human Samples
Abstract
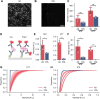
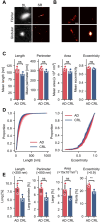
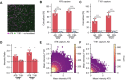
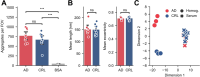
Hyperphosphorylation and aggregation of the protein tau play key roles in the development of Alzheimer's disease (AD). While the molecular structure of the filamentous tau aggregates has been determined to atomic resolution, there is far less information available about the smaller, soluble aggregates, which are believed to be more toxic. Traditional techniques are limited to bulk measures and struggle to identify individual aggregates in complex biological samples. To address this, we developed a novel single-molecule pull-down-based assay (MAPTau) to detect and characterize individual tau aggregates in AD and control post-mortem brain and biofluids. Using MAPTau, we report the quantity, as well as the size and circularity of tau aggregates measured using super-resolution microscopy, revealing AD-specific differences in tau aggregate morphology. By adapting MAPTau to detect multiple phosphorylation markers in individual aggregates using two-color coincidence detection, we derived compositional profiles of the individual aggregates. We find an AD-specific phosphorylation profile of tau aggregates with more than 80 % containing multiple phosphorylations, compared to 5 % in age-matched non-AD controls. Our results show that MAPTau is able to identify disease-specific subpopulations of tau aggregates phosphorylated at different sites, that are invisible to other methods and enable the study of disease mechanisms and diagnosis.
Keywords: fluorescence microscopy; neurodegenerative disease; protein aggregation; proteins; single-molecule studies.
© 2024 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

