Intrinsic apoptosis is evolutionarily divergent among metazoans
- PMID: 38525035
- PMCID: PMC10959488
- DOI: 10.1093/evlett/qrad057
Intrinsic apoptosis is evolutionarily divergent among metazoans
Abstract
Apoptosis is regulated cell death that depends on caspases. A specific initiator caspase is involved upstream of each apoptotic signaling pathway. Characterized in nematode, fly, and mammals, intrinsic apoptosis is considered to be ancestral, conserved among animals, and depends on shared initiators: caspase-9, Apaf-1 and Bcl-2. However, the biochemical role of mitochondria, the pivotal function of cytochrome c and the modality of caspase activation remain highly heterogeneous and hide profound molecular divergence among apoptotic pathways in animals. Uncovering the phylogenetic history of apoptotic actors, especially caspases, is crucial to shed light on the evolutionary history of intrinsic apoptosis. Here, we demonstrate with phylogenetic analyses that caspase-9, the fundamental key of intrinsic apoptosis, is deuterostome-specific, while caspase-2 is ancestral to bilaterians. Our analysis of Bcl-2 and Apaf-1 confirms heterogeneity in functional organization of apoptotic pathways in animals. Our results support emergence of distinct intrinsic apoptotic pathways during metazoan evolution.
Keywords: cell death evolution; initiator caspases; intrinsic apoptosis; phylogeny.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Society for the Study of Evolution (SSE) and European Society for Evolutionary Biology (ESEN).
Figures
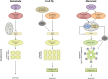

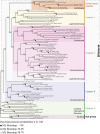
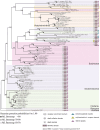


References
-
- An, H. -K., Chung, K. M., Park, H., Hong, J., Gim, J. -E., Choi, H., Lee, Y. W., Choi, J., Mun, J. Y., & Yu, S. -W. (2020). CASP9 (caspase 9) is essential for autophagosome maturation through regulation of mitochondrial homeostasis. Autophagy, 16(9), 1598–1617. 10.1080/15548627.2019.1695398 - DOI - PMC - PubMed
-
- Aubrey, B. J., Janic, A., Chen, Y., Chang, C., Lieschke, E. C., Diepstraten, S. T., Kueh, A. J., Bernardini, J. P., Dewson, G., O’Reilly, L. A., Whitehead, L., Voss, A. K., Smyth, G. K., Strasser, A., & Kelly, G. L. (2018). Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes & Development, 32(21-22), 1420–1429. 10.1101/gad.314286.118 - DOI - PMC - PubMed

