Aethionema arabicum dimorphic seed trait resetting during transition to seedlings
- PMID: 38525145
- PMCID: PMC10957558
- DOI: 10.3389/fpls.2024.1358312
Aethionema arabicum dimorphic seed trait resetting during transition to seedlings
Abstract
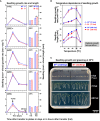
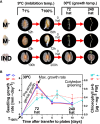
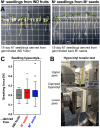
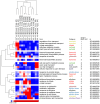
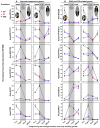
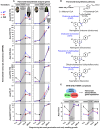
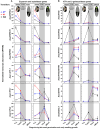
The transition from germinating seeds to emerging seedlings is one of the most vulnerable plant life cycle stages. Heteromorphic diaspores (seed and fruit dispersal units) are an adaptive bet-hedging strategy to cope with spatiotemporally variable environments. While the roles and mechanisms of seedling traits have been studied in monomorphic species, which produce one type of diaspore, very little is known about seedlings in heteromorphic species. Using the dimorphic diaspore model Aethionema arabicum (Brassicaceae), we identified contrasting mechanisms in the germination responses to different temperatures of the mucilaginous seeds (M+ seed morphs), the dispersed indehiscent fruits (IND fruit morphs), and the bare non-mucilaginous M- seeds obtained from IND fruits by pericarp (fruit coat) removal. What follows the completion of germination is the pre-emergence seedling growth phase, which we investigated by comparative growth assays of early seedlings derived from the M+ seeds, bare M- seeds, and IND fruits. The dimorphic seedlings derived from M+ and M- seeds did not differ in their responses to ambient temperature and water potential. The phenotype of seedlings derived from IND fruits differed in that they had bent hypocotyls and their shoot and root growth was slower, but the biomechanical hypocotyl properties of 15-day-old seedlings did not differ between seedlings derived from germinated M+ seeds, M- seeds, or IND fruits. Comparison of the transcriptomes of the natural dimorphic diaspores, M+ seeds and IND fruits, identified 2,682 differentially expressed genes (DEGs) during late germination. During the subsequent 3 days of seedling pre-emergence growth, the number of DEGs was reduced 10-fold to 277 root DEGs and 16-fold to 164 shoot DEGs. Among the DEGs in early seedlings were hormonal regulators, in particular for auxin, ethylene, and gibberellins. Furthermore, DEGs were identified for water and ion transporters, nitrate transporter and assimilation enzymes, and cell wall remodeling protein genes encoding enzymes targeting xyloglucan and pectin. We conclude that the transcriptomes of seedlings derived from the dimorphic diaspores, M+ seeds and IND fruits, undergo transcriptional resetting during the post-germination pre-emergence growth transition phase from germinated diaspores to growing seedlings.
Keywords: bet-hedging strategy; diaspore dimorphism; fruit and seed heteromorphism; pericarp-imposed dormancy; pre-emergence growth; seed seedling transition; seedling stress resilience; transcriptome resetting.
Copyright © 2024 Arshad, Steinbrecher, Wilhelmsson, Fernandez-Pozo, Pérez, Mérai, Rensing, Chandler and Leubner-Metzger.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








Similar articles
-
Dispersal biophysics and adaptive significance of dimorphic diaspores in the annual Aethionema arabicum (Brassicaceae).New Phytol. 2019 Feb;221(3):1434-1446. doi: 10.1111/nph.15490. Epub 2018 Oct 25. New Phytol. 2019. PMID: 30230555 Free PMC article.
-
The dimorphic diaspore model Aethionema arabicum (Brassicaceae): Distinct molecular and morphological control of responses to parental and germination temperatures.Plant Cell. 2024 Jul 2;36(7):2465-2490. doi: 10.1093/plcell/koae085. Plant Cell. 2024. PMID: 38513609 Free PMC article.
-
A tale of two morphs: developmental patterns and mechanisms of seed coat differentiation in the dimorphic diaspore model Aethionema arabicum (Brassicaceae).Plant J. 2021 Jul;107(1):166-181. doi: 10.1111/tpj.15283. Epub 2021 May 18. Plant J. 2021. PMID: 33945185
-
Seed germination variability: why do genetically identical seeds not germinate at the same time?J Exp Bot. 2023 Jun 27;74(12):3462-3475. doi: 10.1093/jxb/erad101. J Exp Bot. 2023. PMID: 36946595 Review.
-
Exploring fine tuning between phytohormones and ROS signaling cascade in regulation of seed dormancy, germination and seedling development.Plant Physiol Biochem. 2024 Feb;207:108352. doi: 10.1016/j.plaphy.2024.108352. Epub 2024 Jan 14. Plant Physiol Biochem. 2024. PMID: 38266558 Review.
References
-
- Alexa A., Rahnenfuhrer J. (2021). topGO: Enrichment analysis for Gene Ontology. R package version 2.44.0.
-
- Arshad W., Lenser T., Wilhelmsson P. K. I., Chandler J. O., Steinbrecher T., Marone F., et al. . (2021). A tale of two morphs: developmental patterns and mechanisms of seed coat differentiation in the dimorphic diaspore model Aethionema arabicum (Brassicaceae). Plant J. 107, 166–181. doi: 10.1111/tpj.15283 - DOI - PubMed
-
- Arshad W., Marone F., Collinson M. E., Leubner-Metzger G., Steinbrecher T. (2020). Fracture of the dimorphic fruits of Aethionema arabicum (Brassicaceae). Botany 98, 65–75. doi: 10.1139/cjb-2019-0014 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

