Huayu Qutan Recipe promotes lipophagy and cholesterol efflux through the mTORC1/TFEB/ABCA1-SCARB1 signal axis
- PMID: 38526033
- PMCID: PMC10962127
- DOI: 10.1111/jcmm.18257
Huayu Qutan Recipe promotes lipophagy and cholesterol efflux through the mTORC1/TFEB/ABCA1-SCARB1 signal axis
Abstract
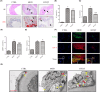
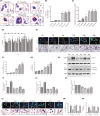
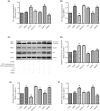
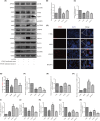
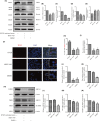
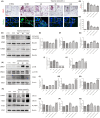
This study aims to investigate the mechanism of the anti-atherosclerosis effect of Huayu Qutan Recipe (HYQT) on the inhibition of foam cell formation. In vivo, the mice were randomly divided into three groups: CTRL group, MOD group and HYQT group. The HYQT group received HYQT oral administration twice a day (20.54 g/kg/d), and the plaque formation in ApoE-/- mice was observed using haematoxylin-eosin (HE) staining and oil red O (ORO) staining. The co-localization of aortic macrophages and lipid droplets (LDs) was examined using fluorescent labelling of CD11b and BODIPY fluorescence probe. In vitro, RAW 264.7 cells were exposed to 50 μg/mL ox-LDL for 48 h and then treated with HYQT for 24 h. The accumulation of LDs was evaluated using ORO and BODIPY. Cell viability was assessed using the CCK-8 assay. The co-localization of LC3b and BODIPY was detected via immunofluorescence and fluorescence probe. LysoTracker Red and BODIPY 493/503 were used as markers for lysosomes and LDs, respectively. Autophagosome formation were observed via transmission electron microscopy. The levels of LC3A/B II/LC3A/B I, p-mTOR/mTOR, p-4EBP1/4EBP1, p-P70S6K/P70S6K and TFEB protein level were examined via western blotting, while SQSTM1/p62, Beclin1, ABCA1, ABCG1 and SCARB1 were examined via qRT-PCR and western blotting. The nuclear translocation of TFEB was detected using immunofluorescence. The components of HYQT medicated serum were determined using Q-Orbitrap high-resolution MS analysis. Molecular docking was employed to identify the components of HYQT medicated serum responsible for the mTOR signalling pathway. The mechanism of taurine was illustrated. HYQT has a remarkable effect on atherosclerotic plaque formation and blood lipid level in ApoE-/- mice. HYQT decreased the co-localization of CD11b and BODIPY. HYQT (10% medicated serum) reduced the LDs accumulation in RAW 264.7 cells. HYQT and RAPA (rapamycin, a mTOR inhibitor) could promote cholesterol efflux, while chloroquine (CQ, an autophagy inhibitor) weakened the effect of HYQT. Moreover, MHY1485 (a mTOR agonist) also mitigated the effects of HYQT by reduced cholesterol efflux. qRT-PCR and WB results suggested that HYQT improved the expression of the proteins ABCA1, ABCG1 and SCARB1.HYQT regulates ABCA1 and SCARB1 protein depending on the mTORC1/TFEB signalling pathway. However, the activation of ABCG1 does not depend on this pathway. Q-Orbitrap high-resolution MS analysis results demonstrated that seven core compounds have good binding ability to the mTOR protein. Taurine may play an important role in the mechanism regulation. HYQT may reduce cardiovascular risk by promoting cholesterol efflux and degrading macrophage-derived foam cell formation. It has been observed that HYQT and ox-LDL regulate lipophagy through the mTOR/TFEB signalling pathway, rather than the mTOR/4EBP1/P70S6K pathway. Additionally, HYQT is found to regulate cholesterol efflux through the mTORC1/TFEB/ABCA1-SCARB1 signal axis, while taurine plays a significant role in lipophagy.
Keywords: autophagy; lipid metabolism; macrophage; ox‐LDL.
© 2024 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The author reports no conflicts of interest in this work.
Figures



References
-
- World Health Organization [homepage on the Internet] . Cardiovascular diseases (CVDs) Fact Sheet. 2017. Available from: https://www.who.int/zh/news‐room/fact‐sheets/detail/cardiovascular‐disea... Accessed Mar 19, 2023.
Publication types
MeSH terms
Substances
Grants and funding
- ZYZX1907/Open Fund of Key Laboratory of Theory and Application of Viscera- manifestation theory in Traditional Chinese Medicine, Ministry of Education, Liaoning University of TCM
- L201722/Science and Technology Research of Education Department of Liaoning Province
- LJKQZ2021064/Youth Project of Basic Scientific Research Project of Education Department of Liaoning Province
- YM202109/The "Seedling Raising Project" of The Affiliated Hospital of Liaoning University of Chinese Medicine
- L202048/Science and Technology Research Project of Education Department of Liaoning Province
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

