Characterization of individual bile acids in vivo utilizing a novel low bile acid mouse model
- PMID: 38526215
- PMCID: PMC12104498
- DOI: 10.1093/toxsci/kfae029
Characterization of individual bile acids in vivo utilizing a novel low bile acid mouse model
Abstract
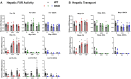
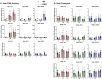
Bile acids (BAs) are signaling molecules synthesized in the liver initially by CYP7A1 and CYP27A1 in the classical and alternative pathways, respectively. BAs are essential for cholesterol clearance, intestinal absorption of lipids, and endogenous modulators of farnesoid x receptor (FXR). FXR is critical in maintaining BA homeostasis and gut-liver crosstalk. Complex reactions in vivo and the lack of suitable animal models impede our understanding of the functions of individual BAs. In this study, we characterized the in vivo effects of three-day feeding of cholic acid (CA), deoxycholic acid (DCA), or ursodeoxycholic acid (UDCA) at physiological/non-hepatotoxic concentrations in a novel low-BA mouse model (Cyp7a1-/-/Cyp27a1-/-, DKO). Liver injury, BA levels and composition and BA signaling by the FXR-fibroblast growth factor 15 (FGF15) axis were determined. Overall, higher basal inflammation and altered lipid metabolism in DKO mice might be associated with low BAs. CA, DCA, and UDCA feeding activated FXR signals with tissue specificity. Dietary CA and DCA similarly altered tissue BA profiles to be less hydrophobic, while UDCA promoted a more hydrophobic tissue BA pool with the profiles shifted toward non-12α-OH BAs and secondary BAs. However, UDCA did not offer any overt protective effects as expected. These findings allow us to determine the precise effects of individual BAs in vivo on BA-FXR signaling and overall BA homeostasis in liver physiology and pathologies.
Keywords: bile acids; liver; nuclear receptors.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society of Toxicology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures









Similar articles
-
Effects of therapeutically approved individual bile acids on the development of metabolic dysfunction-associated steatohepatitis a low bile acid mouse model.Toxicol Sci. 2024 Dec 1;202(2):179-195. doi: 10.1093/toxsci/kfae110. Toxicol Sci. 2024. PMID: 39302723 Free PMC article.
-
Individual bile acids have differential effects on bile acid signaling in mice.Toxicol Appl Pharmacol. 2015 Feb 15;283(1):57-64. doi: 10.1016/j.taap.2014.12.005. Epub 2015 Jan 9. Toxicol Appl Pharmacol. 2015. PMID: 25582706 Free PMC article.
-
Bile Acid Homeostasis in a Cholesterol 7α-Hydroxylase and Sterol 27-Hydroxylase Double Knockout Mouse Model.Hepatology. 2019 Jul;70(1):389-402. doi: 10.1002/hep.30612. Epub 2019 Apr 25. Hepatology. 2019. PMID: 30864232 Free PMC article.
-
The gut-liver axis in hepatocarcinoma: a focus on the nuclear receptor FXR and the enterokine FGF19.Curr Opin Pharmacol. 2018 Dec;43:93-98. doi: 10.1016/j.coph.2018.08.005. Epub 2018 Sep 14. Curr Opin Pharmacol. 2018. PMID: 30223181 Review.
-
Bile acid nuclear receptor FXR and digestive system diseases.Acta Pharm Sin B. 2015 Mar;5(2):135-44. doi: 10.1016/j.apsb.2015.01.004. Epub 2015 Feb 25. Acta Pharm Sin B. 2015. PMID: 26579439 Free PMC article. Review.
Cited by
-
Adrenal High-Expressional CYP27A1 Mediates Bile Acid Increase and Functional Impairment in Adult Male Offspring by Prenatal Dexamethasone Exposure.Adv Sci (Weinh). 2025 Apr;12(14):e2413299. doi: 10.1002/advs.202413299. Epub 2025 Feb 14. Adv Sci (Weinh). 2025. PMID: 39950753 Free PMC article.
-
Crosstalk between bile acids and gut microbiota: a potential target for precancerous lesions of gastric cancer.Front Pharmacol. 2025 Mar 13;16:1533141. doi: 10.3389/fphar.2025.1533141. eCollection 2025. Front Pharmacol. 2025. PMID: 40183085 Free PMC article. Review.
-
Deletion of hepatic FXR leads to more severe MASH development in female mice.Hepatol Commun. 2025 May 16;9(6):e0693. doi: 10.1097/HC9.0000000000000693. eCollection 2025 Jun 1. Hepatol Commun. 2025. PMID: 40377496 Free PMC article.
-
Effects of therapeutically approved individual bile acids on the development of metabolic dysfunction-associated steatohepatitis a low bile acid mouse model.Toxicol Sci. 2024 Dec 1;202(2):179-195. doi: 10.1093/toxsci/kfae110. Toxicol Sci. 2024. PMID: 39302723 Free PMC article.
-
Humanized monoacylglycerol acyltransferase 2 mice develop metabolic dysfunction-associated steatohepatitis.J Lipid Res. 2024 Dec;65(12):100695. doi: 10.1016/j.jlr.2024.100695. Epub 2024 Nov 5. J Lipid Res. 2024. PMID: 39505262 Free PMC article.
References
-
- Barrasa J. I., Olmo N., Lizarbe M. A., Turnay J. (2013). Bile acids in the Colon, from healthy to cytotoxic molecules. Toxicol. In Vitro. 27, 964–977. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

