Phenome-wide Mendelian randomisation analysis of 378,142 cases reveals risk factors for eight common cancers
- PMID: 38527997
- PMCID: PMC10963765
- DOI: 10.1038/s41467-024-46927-z
Phenome-wide Mendelian randomisation analysis of 378,142 cases reveals risk factors for eight common cancers
Abstract
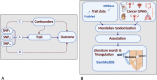
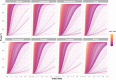
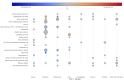
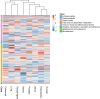
For many cancers there are only a few well-established risk factors. Here, we use summary data from genome-wide association studies (GWAS) in a Mendelian randomisation (MR) phenome-wide association study (PheWAS) to identify potentially causal relationships for over 3,000 traits. Our outcome datasets comprise 378,142 cases across breast, prostate, colorectal, lung, endometrial, oesophageal, renal, and ovarian cancers, as well as 485,715 controls. We complement this analysis by systematically mining the literature space for supporting evidence. In addition to providing supporting evidence for well-established risk factors (smoking, alcohol, obesity, lack of physical activity), we also find sex steroid hormones, plasma lipids, and telomere length as determinants of cancer risk. A number of the molecular factors we identify may prove to be potential biomarkers. Our analysis, which highlights aetiological similarities and differences in common cancers, should aid public health prevention strategies to reduce cancer burden. We provide a R/Shiny app to visualise findings.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Risk factors for eight common cancers revealed from a phenome-wide Mendelian randomisation analysis of 378,142 cases and 485,715 controls.Res Sq [Preprint]. 2023 Mar 17:rs.3.rs-2587058. doi: 10.21203/rs.3.rs-2587058/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Mar 25;15(1):2637. doi: 10.1038/s41467-024-46927-z. PMID: 36993383 Free PMC article. Updated. Preprint.
-
Risk factors for eight common cancers revealed from a phenome-wide Mendelian randomisation analysis of 378,142 cases and 485,715 controls.medRxiv [Preprint]. 2023 Apr 6:2023.02.15.23285952. doi: 10.1101/2023.02.15.23285952. medRxiv. 2023. Update in: Nat Commun. 2024 Mar 25;15(1):2637. doi: 10.1038/s41467-024-46927-z. PMID: 37066289 Free PMC article. Updated. Preprint.
References
-
- International Agency for Research on Cancer Website. https://www.iarc.who.int. World Cancer Report 2008. (International Agency for Research on Cancer, 2008). Date accessed: 02/01/2023.
-
- Stephen, B. & Simon, G. T. Mendelian randomization: Methods for using genetic variants in causal estimation. Biometrics vol. 73 356–356 (CRC Press, Boca Raton, 2021).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

