DNA methylation profiling in Kabuki syndrome: reclassification of germline KMT2D VUS and sensitivity in validating postzygotic mosaicism
- PMID: 38528056
- PMCID: PMC11220151
- DOI: 10.1038/s41431-024-01597-9
DNA methylation profiling in Kabuki syndrome: reclassification of germline KMT2D VUS and sensitivity in validating postzygotic mosaicism
Abstract
Autosomal dominant Kabuki syndrome (KS) is a rare multiple congenital anomalies/neurodevelopmental disorder caused by heterozygous inactivating variants or structural rearrangements of the lysine-specific methyltransferase 2D (KMT2D) gene. While it is often recognizable due to a distinctive gestalt, the disorder is clinically variable, and a phenotypic scoring system has been introduced to help clinicians to reach a clinical diagnosis. The phenotype, however, can be less pronounced in some patients, including those carrying postzygotic mutations. The full spectrum of pathogenic variation in KMT2D has not fully been characterized, which may hamper the clinical classification of a portion of these variants. DNA methylation (DNAm) profiling has successfully been used as a tool to classify variants in genes associated with several neurodevelopmental disorders, including KS. In this work, we applied a KS-specific DNAm signature in a cohort of 13 individuals with KMT2D VUS and clinical features suggestive or overlapping with KS. We succeeded in correctly classifying all the tested individuals, confirming diagnosis for three subjects and rejecting the pathogenic role of 10 VUS in the context of KS. In the latter group, exome sequencing allowed to identify the genetic cause underlying the disorder in three subjects. By testing five individuals with postzygotic pathogenic KMT2D variants, we also provide evidence that DNAm profiling has power to recognize pathogenic variants at different levels of mosaicism, identifying 15% as the minimum threshold for which DNAm profiling can be applied as an informative diagnostic tool in KS mosaics.
© 2024. The Author(s), under exclusive licence to European Society of Human Genetics.
Conflict of interest statement
The authors declare no competing interests.
Figures
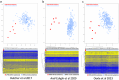
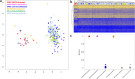

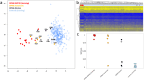
Similar articles
-
Pathogenic variants in KMT2C result in a neurodevelopmental disorder distinct from Kleefstra and Kabuki syndromes.Am J Hum Genet. 2024 Aug 8;111(8):1626-1642. doi: 10.1016/j.ajhg.2024.06.009. Epub 2024 Jul 15. Am J Hum Genet. 2024. PMID: 39013459 Free PMC article.
-
Clinical Characteristics of Patients With Kabuki Syndrome at a Single Tertiary Children's Hospital.Am J Med Genet A. 2025 Jun;197(6):e64003. doi: 10.1002/ajmg.a.64003. Epub 2025 Feb 12. Am J Med Genet A. 2025. PMID: 39936499
-
An HNRNPK-specific DNA methylation signature makes sense of missense variants and expands the phenotypic spectrum of Au-Kline syndrome.Am J Hum Genet. 2022 Oct 6;109(10):1867-1884. doi: 10.1016/j.ajhg.2022.08.014. Epub 2022 Sep 20. Am J Hum Genet. 2022. PMID: 36130591 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Kabuki syndrome: a Chinese case series and systematic review of the spectrum of mutations.BMC Med Genet. 2015 Apr 21;16:26. doi: 10.1186/s12881-015-0171-4. BMC Med Genet. 2015. PMID: 25896430 Free PMC article.
Cited by
-
Hitting the heights with CiteScore.Eur J Hum Genet. 2024 Jul;32(7):743-744. doi: 10.1038/s41431-024-01651-6. Eur J Hum Genet. 2024. PMID: 38956180 Free PMC article. No abstract available.
-
Expanding Upon Genomics in Rare Diseases: Epigenomic Insights.Int J Mol Sci. 2024 Dec 27;26(1):135. doi: 10.3390/ijms26010135. Int J Mol Sci. 2024. PMID: 39795993 Free PMC article. Review.
-
Biallelic loss-of-function variants in ZNF142 are associated with a robust DNA methylation signature affecting a limited number of genomic loci.Eur J Hum Genet. 2025 Jul;33(7):896-903. doi: 10.1038/s41431-025-01876-z. Epub 2025 May 23. Eur J Hum Genet. 2025. PMID: 40410387
-
The Arg99Gln Substitution in HNRNPC Is Associated with a Distinctive Clinical Phenotype Characterized by Facial Dysmorphism and Ocular and Cochlear Anomalies.Genes (Basel). 2025 Feb 1;16(2):176. doi: 10.3390/genes16020176. Genes (Basel). 2025. PMID: 40004505 Free PMC article.
References
-
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. - PubMed
-
- Niikawa N, Matsuura N, Fukushima Y, Ohsawa T, Kajii T. Kabuki make-up syndrome: a syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J Pediatr. 1981;99:565–9. - PubMed
-
- Adam MP, Hudgins L, Hannibal M. Kabuki syndrome. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, et al. editors. GeneReviews [Internet]. Seattle (WA): University of Washington 2011; p. 1993–2023. (updated 2022).
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 5x1000_2019/Ministero della Salute (Ministry of Health, Italy)
- RCR-2022-23682289/Ministero della Salute (Ministry of Health, Italy)
- Current Research Funds/Ministero della Salute (Ministry of Health, Italy)
- FOE_2020/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

