rs822336 binding to C/EBPβ and NFIC modulates induction of PD-L1 expression and predicts anti-PD-1/PD-L1 therapy in advanced NSCLC
- PMID: 38528526
- PMCID: PMC10962156
- DOI: 10.1186/s12943-024-01976-2
rs822336 binding to C/EBPβ and NFIC modulates induction of PD-L1 expression and predicts anti-PD-1/PD-L1 therapy in advanced NSCLC
Abstract
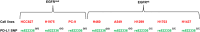
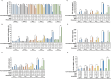
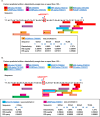
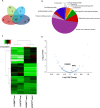
Efficient predictive biomarkers are needed for immune checkpoint inhibitor (ICI)-based immunotherapy in non-small cell lung cancer (NSCLC). Testing the predictive value of single nucleotide polymorphisms (SNPs) in programmed cell death 1 (PD-1) or its ligand 1 (PD-L1) has shown contrasting results. Here, we aim to validate the predictive value of PD-L1 SNPs in advanced NSCLC patients treated with ICIs as well as to define the molecular mechanisms underlying the role of the identified SNP candidate. rs822336 efficiently predicted response to anti-PD-1/PD-L1 immunotherapy in advanced non-oncogene addicted NSCLC patients as compared to rs2282055 and rs4143815. rs822336 mapped to the promoter/enhancer region of PD-L1, differentially affecting the induction of PD-L1 expression in human NSCLC cell lines as well as their susceptibility to HLA class I antigen matched PBMCs incubated with anti-PD-1 monoclonal antibody nivolumab. The induction of PD-L1 expression by rs822336 was mediated by a competitive allele-specificity binding of two identified transcription factors: C/EBPβ and NFIC. As a result, silencing of C/EBPβ and NFIC differentially regulated the induction of PD-L1 expression in human NSCLC cell lines carrying different rs822336 genotypes. Analysis by binding microarray further validated the competitive allele-specificity binding of C/EBPβ and NFIC to PD-L1 promoter/enhancer region based on rs822336 genotype in human NSCLC cell lines. These findings have high clinical relevance since identify rs822336 and induction of PD-L1 expression as novel biomarkers for predicting anti-PD-1/PD-L1-based immunotherapy in advanced NSCLC patients.
Keywords: Immunotherapy; NSCLC; PD-1; PD-L1; Predictive biomarker; SNP; rs822336.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Scagliotti GV, Parikh P, von Pawel J, Biesma B, Vansteenkiste J, Manegold C, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:3543–51. doi: 10.1200/JCO.2007.15.0375. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

