The Dual Role of the NFATc2/galectin-9 Axis in Modulating Tumor-Initiating Cell Phenotypes and Immune Suppression in Lung Adenocarcinoma
- PMID: 38528665
- PMCID: PMC11132051
- DOI: 10.1002/advs.202306059
The Dual Role of the NFATc2/galectin-9 Axis in Modulating Tumor-Initiating Cell Phenotypes and Immune Suppression in Lung Adenocarcinoma
Abstract
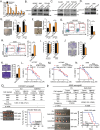
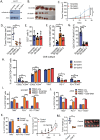
Tumor-initiating cells (TICs) resilience and an immunosuppressive microenvironment are aggressive oncogenic phenotypes that contribute to unsatisfactory long-term outcomes in lung adenocarcinoma (LUAD) patients. The molecular mechanisms mediating the interaction between TICs and immune tolerance have not been elucidated. The role of Galectin-9 in oncogenesis and immunosuppressive microenvironment is still unknown. This study explored the potential role of galectin-9 in TIC regulation and immune modulation in LUAD. The results show that galectin-9 supports TIC properties in LUAD. Co-culture of patient-derived organoids and matched peripheral blood mononuclear cells showed that tumor-secreted galectin-9 suppressed T cell cytotoxicity and induced regulatory T cells (Tregs). Clinically, galectin-9 is upregulated in human LUAD. High expression of galectin-9 predicted poor recurrence-free survival and correlated with high levels of Treg infiltration. LGALS9, the gene encoding galectin-9, is found to be transcriptionally regulated by the nuclear factor of activated T cells 2 (NFATc2), a previously reported TIC regulator, via in silico prediction and luciferase reporter assays. Overall, the results suggest that the NFATc2/galectin-9 axis plays a dual role in TIC regulation and immune suppression.
Keywords: NFATc2; galectin‐9; immune; lung adenocarcinoma; tumor‐initiating cells.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures





References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J. Clin. 2021, 71, 209. - PubMed
-
- Kreso A., Dick J. E., Cell Stem Cell 2014, 14, 275. - PubMed
-
- Clarke M. F., N. Engl. J. Med. 2019, 380, 2237. - PubMed
-
- Garon E. B., Rizvi N. A., Hui R., Leighl N., Balmanoukian A. S., Eder J. P., Patnaik A., Aggarwal C., Gubens M., Horn L., Carcereny E., Ahn M.‐J., Felip E., Lee J.‐S., Hellmann M. D., Hamid O., Goldman J. W., Soria J.‐C., Dolled‐Filhart M., Rutledge R. Z., Zhang J., Lunceford J. K., Rangwala R., Lubiniecki G. M., Roach C., Emancipator K., Gandhi L., N. Engl. J. Med. 2015, 372, 2018. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
