Pseudomonas putida as a platform for medium-chain length α,ω-diol production: Opportunities and challenges
- PMID: 38528784
- PMCID: PMC10963910
- DOI: 10.1111/1751-7915.14423
Pseudomonas putida as a platform for medium-chain length α,ω-diol production: Opportunities and challenges
Abstract
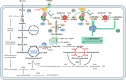
Medium-chain-length α,ω-diols (mcl-diols) play an important role in polymer production, traditionally depending on energy-intensive chemical processes. Microbial cell factories offer an alternative, but conventional strains like Escherichia coli and Saccharomyces cerevisiae face challenges in mcl-diol production due to the toxicity of intermediates such as alcohols and acids. Metabolic engineering and synthetic biology enable the engineering of non-model strains for such purposes with P. putida emerging as a promising microbial platform. This study reviews the advancement in diol production using P. putida and proposes a four-module approach for the sustainable production of diols. Despite progress, challenges persist, and this study discusses current obstacles and future opportunities for leveraging P. putida as a microbial cell factory for mcl-diol production. Furthermore, this study highlights the potential of using P. putida as an efficient chassis for diol synthesis.
© 2024 The Authors. Microbial Biotechnology published by Applied Microbiology International and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Arkatkar, A. , Juwarkar, A.A. , Bhaduri, S. , Uppara, P.V. & Doble, M. (2010) Growth of pseudomonas and bacillus biofilms on pretreated polypropylene surface. International Biodeterioration and Biodegradation, 64, 530–536.
-
- Balasubramanian, V. , Natarajan, K. , Hemambika, B. , Ramesh, N. , Sumathi, C.S. , Kottaimuthu, R. et al. (2010) High‐density polyethylene (HDPE)‐degrading potential bacteria from marine ecosystem of gulf of Mannar, India. Letters in Applied Microbiology, 51, 205–211. - PubMed
-
- Batianis, C. , van Rosmalen, R.P. , Major, M. , van Ee, C. , Kasiotakis, A. , Weusthuis, R.A. et al. (2023) A tunable metabolic valve for precise growth control and increased product formation in Pseudomonas putida . Metabolic Engineering, 75, 47–57. - PubMed
-
- Belda, E. , van Heck, R.G.A. , José Lopez‐Sanchez, M. , Cruveiller, S. , Barbe, V. , Fraser, C. et al. (2016) The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environmental Microbiology, 18, 3403–3424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

