Dysregulation of LINC00324 promotes poor prognosis in patients with glioma
- PMID: 38530810
- PMCID: PMC10965094
- DOI: 10.1371/journal.pone.0298055
Dysregulation of LINC00324 promotes poor prognosis in patients with glioma
Abstract
Background: LINC00324 is a long-stranded non-coding RNA, which is aberrantly expressed in various cancers and is associated with poor prognosis and clinical features. It involves multiple oncogenic molecular pathways affecting cell proliferation, migration, invasion, and apoptosis. However, the expression, function, and mechanism of LINC00324 in glioma have not been reported.
Material and methods: We assessed the expression of LINC00324 of LINC00324 in glioma patients based on data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) to identify pathways involved in LINC00324-related glioma pathogenesis.
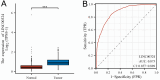
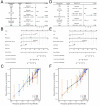
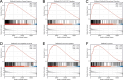
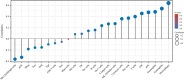
Results: Based on our findings, we observed differential expression of LINC00324 between tumor and normal tissues in glioma patients. Our analysis of overall survival (OS) and disease-specific survival (DSS) indicated that glioma patients with high LINC00324 expression had a poorer prognosis compared to those with low LINC00324 expression. By integrating clinical data and genetic signatures from TCGA patients, we developed a nomogram to predict OS and DSS in glioma patients. Gene set enrichment analysis (GSEA) revealed that several pathways, including JAK/STAT3 signaling, epithelial-mesenchymal transition, STAT5 signaling, NF-κB activation, and apoptosis, were differentially enriched in glioma samples with high LINC00324 expression. Furthermore, we observed significant correlations between LINC00324 expression, immune infiltration levels, and expression of immune checkpoint-related genes (HAVCR2: r = 0.627, P = 1.54e-77; CD40: r = 0.604, P = 1.36e-70; ITGB2: r = 0.612, P = 6.33e-7; CX3CL1: r = -0.307, P = 9.24e-17). These findings highlight the potential significance of LINC00324 in glioma progression and suggest avenues for further research and potential therapeutic targets.
Conclusion: Indeed, our results confirm that the LINC00324 signature holds promise as a prognostic predictor in glioma patients. This finding opens up new possibilities for understanding the disease and may offer valuable insights for the development of targeted therapies.
Copyright: © 2024 Jin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
MCM10 as a novel prognostic biomarker and its relevance to immune infiltration in gliomas.Technol Health Care. 2023;31(4):1301-1317. doi: 10.3233/THC-220576. Technol Health Care. 2023. PMID: 36872806
-
LINC00324 accelerates the proliferation and migration of osteosarcoma through regulating WDR66.J Cell Physiol. 2020 Jan;235(1):339-348. doi: 10.1002/jcp.28973. Epub 2019 Jun 21. J Cell Physiol. 2020. PMID: 31225659
-
Selenoprotein GPX1 is a prognostic and chemotherapy-related biomarker for brain lower grade glioma.J Trace Elem Med Biol. 2022 Dec;74:127082. doi: 10.1016/j.jtemb.2022.127082. Epub 2022 Sep 17. J Trace Elem Med Biol. 2022. PMID: 36155420
-
Long noncoding RNA LINC00324 promotes retinoblastoma progression by acting as a competing endogenous RNA for microRNA-769-5p, thereby increasing STAT3 expression.Aging (Albany NY). 2020 May 5;12(9):7729-7746. doi: 10.18632/aging.103075. Epub 2020 May 5. Aging (Albany NY). 2020. PMID: 32369777 Free PMC article.
-
LINC00324 in cancer: Regulatory and therapeutic implications.Front Oncol. 2022 Dec 22;12:1039366. doi: 10.3389/fonc.2022.1039366. eCollection 2022. Front Oncol. 2022. PMID: 36620587 Free PMC article. Review.
Cited by
-
Clinical significance and biological function of PRKCQ-AS1/miR-582-3p expression in LUAD.Hereditas. 2025 Jul 1;162(1):116. doi: 10.1186/s41065-025-00482-9. Hereditas. 2025. PMID: 40597380 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

