Local-scale phylodynamics reveal differential community impact of SARS-CoV-2 in a metropolitan US county
- PMID: 38530853
- PMCID: PMC10997136
- DOI: 10.1371/journal.ppat.1012117
Local-scale phylodynamics reveal differential community impact of SARS-CoV-2 in a metropolitan US county
Abstract
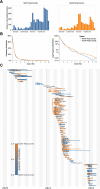
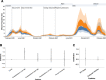
SARS-CoV-2 transmission is largely driven by heterogeneous dynamics at a local scale, leaving local health departments to design interventions with limited information. We analyzed SARS-CoV-2 genomes sampled between February 2020 and March 2022 jointly with epidemiological and cell phone mobility data to investigate fine scale spatiotemporal SARS-CoV-2 transmission dynamics in King County, Washington, a diverse, metropolitan US county. We applied an approximate structured coalescent approach to model transmission within and between North King County and South King County alongside the rate of outside introductions into the county. Our phylodynamic analyses reveal that following stay-at-home orders, the epidemic trajectories of North and South King County began to diverge. We find that South King County consistently had more reported and estimated cases, COVID-19 hospitalizations, and longer persistence of local viral transmission when compared to North King County, where viral importations from outside drove a larger proportion of new cases. Using mobility and demographic data, we also find that South King County experienced a more modest and less sustained reduction in mobility following stay-at-home orders than North King County, while also bearing more socioeconomic inequities that might contribute to a disproportionate burden of SARS-CoV-2 transmission. Overall, our findings suggest a role for local-scale phylodynamics in understanding the heterogeneous transmission landscape.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
ALG reported receiving contract testing from Abbott, Cepheid, Novavax, Pfizer, Janssen and Hologic and research support from Gilead and Merck, outside of the described work. HC reported consulting with Ellume, Pfizer, the Bill & Melinda Gates Foundation, Glaxo Smith Kline, and Merck. She has received research funding from Emergent Ventures, Gates Ventures, Sanofi Pasteur, the Bill & Melinda Gates Foundation, and research support and reagents from Ellume and Cepheid outside of the submitted work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All other authors declare no competing interests.
Figures




Update of
-
Local-Scale phylodynamics reveal differential community impact of SARS-CoV-2 in metropolitan US county.medRxiv [Preprint]. 2022 Dec 16:2022.12.15.22283536. doi: 10.1101/2022.12.15.22283536. medRxiv. 2022. Update in: PLoS Pathog. 2024 Mar 26;20(3):e1012117. doi: 10.1371/journal.ppat.1012117. PMID: 36561171 Free PMC article. Updated. Preprint.
References
-
- Tordoff DM, Greninger AL, Roychoudhury P, Shrestha L, Xie H, Jerome KR, et al.. Phylogenetic estimates of SARS-CoV-2 introductions into Washington State. Lancet Reg Health–Am [Internet]. 2021. Sep 1 [cited 2022 Aug 3];1. Available from: https://www.thelancet.com/journals/lanam/article/PIIS2667-193X(21)00010-... - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

