Neofunctionalization of an OMT cluster dominates polymethoxyflavone biosynthesis associated with the domestication of citrus
- PMID: 38530892
- PMCID: PMC10998556
- DOI: 10.1073/pnas.2321615121
Neofunctionalization of an OMT cluster dominates polymethoxyflavone biosynthesis associated with the domestication of citrus
Abstract
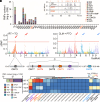
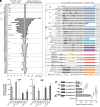
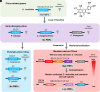
Polymethoxyflavones (PMFs) are a class of abundant specialized metabolites with remarkable anticancer properties in citrus. Multiple methoxy groups in PMFs are derived from methylation modification catalyzed by a series of hydroxylases and O-methyltransferases (OMTs). However, the specific OMTs that catalyze the systematic O-methylation of hydroxyflavones remain largely unknown. Here, we report that PMFs are highly accumulated in wild mandarins and mandarin-derived accessions, while undetectable in early-diverging citrus species and related species. Our results demonstrated that three homologous genes, CreOMT3, CreOMT4, and CreOMT5, are crucial for PMF biosynthesis in citrus, and their encoded methyltransferases exhibit multisite O-methylation activities for hydroxyflavones, producing seven PMFs in vitro and in vivo. Comparative genomic and syntenic analyses indicated that the tandem CreOMT3, CreOMT4, and CreOMT5 may be duplicated from CreOMT6 and contributes to the genetic basis of PMF biosynthesis in the mandarin group through neofunctionalization. We also demonstrated that N17 in CreOMT4 is an essential amino acid residue for C3-, C5-, C6-, and C3'-O-methylation activity and provided a rationale for the functional deficiency of OMT6 to produce PMFs in early-diverging citrus and some domesticated citrus species. A 1,041-bp deletion in the CreOMT4 promoter, which is found in most modern cultivated mandarins, has reduced the PMF content relative to that in wild and early-admixture mandarins. This study provides a framework for reconstructing PMF biosynthetic pathways, which may facilitate the breeding of citrus fruits with enhanced health benefits.
Keywords: O-methyltransferase; citrus; neofunctionalization; polymethoxyflavone; tandemly duplicated gene cluster.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Wang Y., et al. , Polymethoxyflavones from citrus inhibited gastric cancer cell proliferation through inducing apoptosis by upregulating RARβ, both in vitro and in vivo. Food Chem. Toxicol. 146, 111811 (2020). - PubMed
-
- Murakami A., et al. , Inhibitory effect of citrus nobiletin on phorbol ester-induced skin inflammation, oxidative stress, and tumor promotion in mice. Cancer Res. 60, 5059–5066 (2000). - PubMed
-
- Wang H., et al. , Natural Citrus flavanone 5-demethylnobiletin stimulates melanogenesis through the activation of cAMP/CREB pathway in B16F10 cells. Phytomedicine 98, 153941 (2022). - PubMed
-
- Sun Y., et al. , Inhibitory effects of nobiletin and its major metabolites on lung tumorigenesis. Food Funct. 10, 7444–7452 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFD2300600/National Key Research and Development Program of China
- 2021hszd016/Hubei Hongshan Laboratory
- 2023YFD2300600/National Key Research and Development Program of China
- 32272685/National Natural Science Foundation of China
- 2021CFA017/Key project of Hubei Provincial Natural Science Foundation
LinkOut - more resources
Full Text Sources
Miscellaneous

