Mouse Genome Informatics: an integrated knowledgebase system for the laboratory mouse
- PMID: 38531069
- PMCID: PMC11075557
- DOI: 10.1093/genetics/iyae031
Mouse Genome Informatics: an integrated knowledgebase system for the laboratory mouse
Abstract
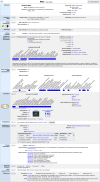
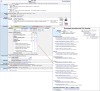
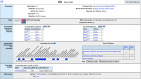
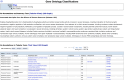
Mouse Genome Informatics (MGI) is a federation of expertly curated information resources designed to support experimental and computational investigations into genetic and genomic aspects of human biology and disease using the laboratory mouse as a model system. The Mouse Genome Database (MGD) and the Gene Expression Database (GXD) are core MGI databases that share data and system architecture. MGI serves as the central community resource of integrated information about mouse genome features, variation, expression, gene function, phenotype, and human disease models acquired from peer-reviewed publications, author submissions, and major bioinformatics resources. To facilitate integration and standardization of data, biocuration scientists annotate using terms from controlled metadata vocabularies and biological ontologies (e.g. Mammalian Phenotype Ontology, Mouse Developmental Anatomy, Disease Ontology, Gene Ontology, etc.), and by applying international community standards for gene, allele, and mouse strain nomenclature. MGI serves basic scientists, translational researchers, and data scientists by providing access to FAIR-compliant data in both human-readable and compute-ready formats. The MGI resource is accessible at https://informatics.jax.org. Here, we present an overview of the core data types represented in MGI and highlight recent enhancements to the resource with a focus on new data and functionality for MGD and GXD.
Keywords: gene expression; genetics; genome informatics; model organism; mouse models; phenotypes.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflicts of interest.
Figures








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

