Multipad agarose plate: a rapid and high-throughput approach for antibiotic susceptibility testing
- PMID: 38531408
- PMCID: PMC10973877
- DOI: 10.1098/rsif.2023.0730
Multipad agarose plate: a rapid and high-throughput approach for antibiotic susceptibility testing
Abstract
We describe a phenotypic antibiotic susceptibility testing (AST) method that can provide an eightfold speed-up in turnaround time compared with the current clinical standard by leveraging advances in microscopy and single-cell imaging. A newly developed growth plate containing 96 agarose pads, termed the multipad agarose plate (MAP), can be assembled at low cost. Pads can be prepared with dilution series of antibiotics. Bacteria are seeded on the pads and automatically imaged using brightfield microscopy, with a fully automated segmentation pipeline quantifying microcolony formation and growth rate. Using a test set of nine antibiotics with very different targets, we demonstrate that accurate minimum inhibitory concentration (MIC) measurements can be performed based on the growth rate of microcolonies within 3 h of incubation with the antibiotic when started from exponential phase. Faster, reliable and high-throughput methods for AST, such as MAP, could improve patient care by expediting treatment initiation and alleviating the burden of antimicrobial resistance.
Keywords: antibiotic susceptibility testing; antimicrobial resistance; imaging bacteria.
Conflict of interest statement
A.D. declares to work for a company that operates in the application areas described in this work, but has no direct conflict of interest.
Figures


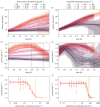
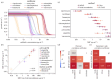
References
-
- WHO. 2020. Antibiotic resistance. See https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed 28 June 2023).
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
