RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy
- PMID: 38531882
- PMCID: PMC10966055
- DOI: 10.1038/s41392-024-01777-5
RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy
Abstract
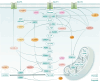
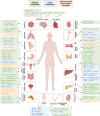
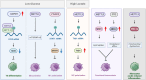
Cellular metabolism is an intricate network satisfying bioenergetic and biosynthesis requirements of cells. Relevant studies have been constantly making inroads in our understanding of pathophysiology, and inspiring development of therapeutics. As a crucial component of epigenetics at post-transcription level, RNA modification significantly determines RNA fates, further affecting various biological processes and cellular phenotypes. To be noted, immunometabolism defines the metabolic alterations occur on immune cells in different stages and immunological contexts. In this review, we characterize the distribution features, modifying mechanisms and biological functions of 8 RNA modifications, including N6-methyladenosine (m6A), N6,2'-O-dimethyladenosine (m6Am), N1-methyladenosine (m1A), 5-methylcytosine (m5C), N4-acetylcytosine (ac4C), N7-methylguanosine (m7G), Pseudouridine (Ψ), adenosine-to-inosine (A-to-I) editing, which are relatively the most studied types. Then regulatory roles of these RNA modification on metabolism in diverse health and disease contexts are comprehensively described, categorized as glucose, lipid, amino acid, and mitochondrial metabolism. And we highlight the regulation of RNA modifications on immunometabolism, further influencing immune responses. Above all, we provide a thorough discussion about clinical implications of RNA modification in metabolism-targeted therapy and immunotherapy, progression of RNA modification-targeted agents, and its potential in RNA-targeted therapeutics. Eventually, we give legitimate perspectives for future researches in this field from methodological requirements, mechanistic insights, to therapeutic applications.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

