Homocysteine metabolites inhibit autophagy by upregulating miR-21-5p, miR-155-5p, miR-216-5p, and miR-320c-3p in human vascular endothelial cells
- PMID: 38531978
- PMCID: PMC10966103
- DOI: 10.1038/s41598-024-57750-3
Homocysteine metabolites inhibit autophagy by upregulating miR-21-5p, miR-155-5p, miR-216-5p, and miR-320c-3p in human vascular endothelial cells
Abstract
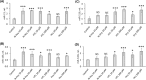
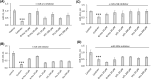
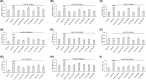
Nutritional and genetic deficiencies in homocysteine (Hcy) metabolism lead to hyperhomocysteinemia (HHcy) and cause endothelial dysfunction, a hallmark of atherosclerosis, which is a major cause of cardiovascular disease (CVD). Impaired autophagy causes the accumulation of damaged proteins and organelles and is associated with CVD. Biochemically, HHcy is characterized by elevated levels of Hcy and its metabolites, Hcy-thiolactone and N-Hcy-protein. However, whether these metabolites can dysregulate mTOR signaling and autophagy in endothelial cells is not known. Here, we examined the influence of Hcy-thiolactone, N-Hcy-protein, and Hcy on autophagy human umbilical vein endothelial cells. We found that treatments with Hcy-thiolactone, N-Hcy-protein, or Hcy significantly downregulated beclin 1 (BECN1), autophagy-related 5 (ATG5), autophagy-related 7 (ATG7), and microtubule-associated protein 1 light chain 3 (LC3) mRNA and protein levels. We also found that these changes were mediated by upregulation by Hcy-thiolactone, N-Hcy-protein, and Hcy of autophagy-targeting microRNA (miR): miR-21, miR-155, miR-216, and miR-320c. The effects of these metabolites on levels of miR targeting autophagy as well as on the levels of BECN1, ATG5, ATG7, and LC3 mRNA and protein were abrogated by treatments with inhibitors of miR-21, miR-155, miR-216, and mir320c. Taken together, our findings show that Hcy metabolites can upregulate miR-21, miR-155, miR-216, and mir320c, which then downregulate autophagy in human endothelial cells, important for vascular homeostasis.
Keywords: Autophagy; Endothelial dysfunction; HUVEC; Homocysteine metabolites; microRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

