A comprehensive computational study to explore promising natural bioactive compounds targeting glycosyltransferase MurG in Escherichia coli for potential drug development
- PMID: 38532068
- PMCID: PMC10966019
- DOI: 10.1038/s41598-024-57702-x
A comprehensive computational study to explore promising natural bioactive compounds targeting glycosyltransferase MurG in Escherichia coli for potential drug development
Abstract
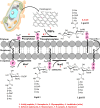
Peptidoglycan is a carbohydrate with a cross-linked structure that protects the cytoplasmic membrane of bacterial cells from damage. The mechanism of peptidoglycan biosynthesis involves the main synthesizing enzyme glycosyltransferase MurG, which is known as a potential target for antibiotic therapy. Many MurG inhibitors have been recognized as MurG targets, but high toxicity and drug-resistant Escherichia coli strains remain the most important problems for further development. In addition, the discovery of selective MurG inhibitors has been limited to the synthesis of peptidoglycan-mimicking compounds. The present study employed drug discovery, such as virtual screening using molecular docking, drug likeness ADMET proprieties predictions, and molecular dynamics (MD) simulation, to identify potential natural products (NPs) for Escherichia coli. We conducted a screening of 30,926 NPs from the NPASS database. Subsequently, 20 of these compounds successfully passed the potency, pharmacokinetic, ADMET screening assays, and their validation was further confirmed through molecular docking. The best three hits and the standard were chosen for further MD simulations up to 400 ns and energy calculations to investigate the stability of the NPs-MurG complexes. The analyses of MD simulations and total binding energies suggested the higher stability of NPC272174. The potential compounds can be further explored in vivo and in vitro for promising novel antibacterial drug discovery.
Keywords: Escherichia coli; Antibacterial; Antibiotics resistance; Molecular dynamics; MurG; Natural products; Virtual screening.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Similar articles
-
E. Coli MurG: a paradigm for a superfamily of glycosyltransferases.Curr Drug Targets Infect Disord. 2001 Aug;1(2):201-13. doi: 10.2174/1568005014606116. Curr Drug Targets Infect Disord. 2001. PMID: 12455415 Review.
-
Screening of potential phytomolecules against MurG as drug target in nosocomial pathogen Pseudomonas aeruginosa: perceptions from computational campaign.J Biomol Struct Dyn. 2024 Jan-Feb;42(1):495-508. doi: 10.1080/07391102.2023.2194005. Epub 2023 Mar 28. J Biomol Struct Dyn. 2024. PMID: 36974974
-
Discovery of novel inhibitors of Mycobacterium tuberculosis MurG: homology modelling, structure based pharmacophore, molecular docking, and molecular dynamics simulations.J Biomol Struct Dyn. 2018 Sep;36(12):3184-3198. doi: 10.1080/07391102.2017.1384398. Epub 2017 Oct 17. J Biomol Struct Dyn. 2018. PMID: 28948866
-
Computer aided ligand based screening for identification of promising molecules against enzymes involved in peptidoglycan biosynthetic pathway from Acinetobacter baumannii.Microb Pathog. 2020 Oct;147:104205. doi: 10.1016/j.micpath.2020.104205. Epub 2020 Apr 28. Microb Pathog. 2020. PMID: 32353580
-
Core Steps of Membrane-Bound Peptidoglycan Biosynthesis: Recent Advances, Insight and Opportunities.Antibiotics (Basel). 2015 Nov 3;4(4):495-520. doi: 10.3390/antibiotics4040495. Antibiotics (Basel). 2015. PMID: 27025638 Free PMC article. Review.
Cited by
-
Mass spectrometry-based metabolomics approaches to interrogate host-microbiome interactions in mammalian systems.Nat Prod Rep. 2025 Jun 16. doi: 10.1039/d5np00021a. Online ahead of print. Nat Prod Rep. 2025. PMID: 40521991 Free PMC article. Review.
-
Machine Learning-Enabled Drug-Induced Toxicity Prediction.Adv Sci (Weinh). 2025 Apr;12(16):e2413405. doi: 10.1002/advs.202413405. Epub 2025 Feb 3. Adv Sci (Weinh). 2025. PMID: 39899688 Free PMC article. Review.
References
-
- World Health Organization. WHO estimates of the global burden of foodborne diseases: foodborne disease burden epidemiology reference group 2007–2015. World HealthOrganization. https://apps.who.int/iris/handle/10665/199350 (2015)
-
- Bhusal B, Yadav B, Dawadi P, Rijal KR, Ghimire P, Banjara MR. Multi-drug resistance, β-lactamases production, and coexistence of bla (NDM-1) and mcr-1 in Escherichia coli clinical isolates from a referral hospital in Kathmandu. Nepal. Microbiol. Insights. 2023;16:1–8. doi: 10.1177/11786361231152220. - DOI - PMC - PubMed
-
- World Health Organization Diarrhoeal Disease. World HealthOrganization. https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease (2017)
-
- Kotloff KL, et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS) Lancet. 2019;7:e568–e584. doi: 10.1016/s2214-109x(19)30076-2. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

