Exosomal long non-coding RNA TRPM2-AS promotes angiogenesis in gallbladder cancer through interacting with PABPC1 to activate NOTCH1 signaling pathway
- PMID: 38532427
- PMCID: PMC10967197
- DOI: 10.1186/s12943-024-01979-z
Exosomal long non-coding RNA TRPM2-AS promotes angiogenesis in gallbladder cancer through interacting with PABPC1 to activate NOTCH1 signaling pathway
Abstract
Background: Abnormal angiogenesis is crucial for gallbladder cancer (GBC) tumor growth and invasion, highlighting the importance of elucidating the mechanisms underlying this process. LncRNA (long non-coding RNA) is widely involved in the malignancy of GBC. However, conclusive evidence confirming the correlation between lncRNAs and angiogenesis in GBC is lacking.
Methods: LncRNA sequencing was performed to identify the differentially expressed lncRNAs. RT-qPCR, western blot, FISH, and immunofluorescence were used to measure TRPM2-AS and NOTCH1 signaling pathway expression in vitro. Mouse xenograft and lung metastasis models were used to evaluate the biological function of TRPM2-AS during angiogenesis in vivo. EDU, transwell, and tube formation assays were used to detect the angiogenic ability of HUVECs. RIP, RAP, RNA pull-down, dual-luciferase reporter system, and mass spectrometry were used to confirm the interaction between TRPM2-AS, IGF2BP2, NUMB, and PABPC1.
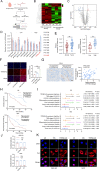
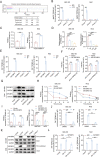
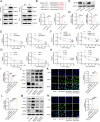
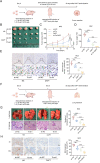
Results: TRPM2-AS was upregulated in GBC tissues and was closely related to angiogenesis and poor prognosis in patients with GBC. The high expression level and stability of TRPM2-AS benefited from m6A modification, which is recognized by IGF2BP2. In terms of exerting pro-angiogenic effects, TRPM2-AS loaded with exosomes transported from GBC cells to HUVECs enhanced PABPC1-mediated NUMB expression inhibition, ultimately promoting the activation of the NOTCH1 signaling pathway. PABPC1 inhibited NUMB mRNA expression through interacting with AGO2 and promoted miR-31-5p and miR-146a-5p-mediated the degradation of NUMB mRNA. The NOTCH signaling pathway inhibitor DAPT inhibited GBC tumor angiogenesis, and TRPM2-AS knockdown enhanced this effect.
Conclusions: TRPM2-AS is a novel and promising biomarker for GBC angiogenesis that promotes angiogenesis by facilitating the activation of the NOTCH1 signaling pathway. Targeting TRPM2-AS opens further opportunities for future GBC treatments.
Keywords: Angiogenesis; Exosome; Gallbladder cancer; IGF2BP2; NOTCH1; PABPC1; TRPM2-AS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

