A hierarchical fusion strategy of deep learning networks for detection and segmentation of hepatocellular carcinoma from computed tomography images
- PMID: 38532511
- PMCID: PMC10964581
- DOI: 10.1186/s40644-024-00686-8
A hierarchical fusion strategy of deep learning networks for detection and segmentation of hepatocellular carcinoma from computed tomography images
Abstract
Background: Automatic segmentation of hepatocellular carcinoma (HCC) on computed tomography (CT) scans is in urgent need to assist diagnosis and radiomics analysis. The aim of this study is to develop a deep learning based network to detect HCC from dynamic CT images.
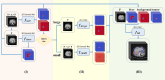
Methods: Dynamic CT images of 595 patients with HCC were used. Tumors in dynamic CT images were labeled by radiologists. Patients were randomly divided into training, validation and test sets in a ratio of 5:2:3, respectively. We developed a hierarchical fusion strategy of deep learning networks (HFS-Net). Global dice, sensitivity, precision and F1-score were used to measure performance of the HFS-Net model.
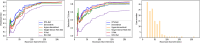
Results: The 2D DenseU-Net using dynamic CT images was more effective for segmenting small tumors, whereas the 2D U-Net using portal venous phase images was more effective for segmenting large tumors. The HFS-Net model performed better, compared with the single-strategy deep learning models in segmenting small and large tumors. In the test set, the HFS-Net model achieved good performance in identifying HCC on dynamic CT images with global dice of 82.8%. The overall sensitivity, precision and F1-score were 84.3%, 75.5% and 79.6% per slice, respectively, and 92.2%, 93.2% and 92.7% per patient, respectively. The sensitivity in tumors < 2 cm, 2-3, 3-5 cm and > 5 cm were 72.7%, 92.9%, 94.2% and 100% per patient, respectively.
Conclusions: The HFS-Net model achieved good performance in the detection and segmentation of HCC from dynamic CT images, which may support radiologic diagnosis and facilitate automatic radiomics analysis.
Keywords: Computed tomography; Deep learning; Detection; Hepatocellular carcinoma; Segmentation.
© 2024. The Author(s).
Conflict of interest statement
YHH has received research grants from Gilead Sciences and Bristol-Meyers Squibb, and honoraria from Abbvie, Gilead Sciences, Bristol-Meyers Squibb, Ono Pharmaceutical, Merck Sharp & Dohme, Eisai, Eli Lilly, Ipsen, and Roche, and has served in an advisory role for Abbvie, Gilead Sciences, Bristol-Meyers Squibb, Ono Pharmaceuticals, Eisai, Eli Lilly, Ipsen, Merck Sharp & Dohme and Roche. The other authors declare no conflicts of interest.
Figures

References
MeSH terms
Grants and funding
- V111C-114, V112C-126/Taipei Veterans General Hospital
- V109E-008-2, V109E-008-2(110)/Taipei Veterans General Hospital
- MOST 108-3011-F-075-001-, 110-2221-E-A49-099-MY3, 111-2740-B-400-002-, 109-2314-B-010-034-MY3/National Science and Technology Council
- 110-2314-B-075-052, 111-2314-B-075-055/National Science and Technology Council
- NSTC 112-2628-B-A49-016-MY3/National Science and Technology Council
LinkOut - more resources
Full Text Sources
Medical

