Transplant of microbiota from Crohn's disease patients to germ-free mice results in colitis
- PMID: 38532703
- PMCID: PMC10978031
- DOI: 10.1080/19490976.2024.2333483
Transplant of microbiota from Crohn's disease patients to germ-free mice results in colitis
Abstract
Although the role of the intestinal microbiota in the pathogenesis of inflammatory bowel disease (IBD) is beyond debate, attempts to verify the causative role of IBD-associated dysbiosis have been limited to reports of promoting the disease in genetically susceptible mice or in chemically induced colitis. We aimed to further test the host response to fecal microbiome transplantation (FMT) from Crohn's disease patients on mucosal homeostasis in ex-germ-free (xGF) mice. We characterized and transferred fecal microbiota from healthy patients and patients with defined Crohn's ileocolitis (CD_L3) to germ-free mice and analyzed the resulting microbial and mucosal homeostasis by 16S profiling, shotgun metagenomics, histology, immunofluorescence (IF) and RNAseq analysis. We observed a markedly reduced engraftment of CD_L3 microbiome compared to healthy control microbiota. FMT from CD_L3 patients did not lead to ileitis but resulted in colitis with features consistent with CD: a discontinued pattern of colitis, more proximal colonic localization, enlarged isolated lymphoid follicles and/or tertiary lymphoid organ neogenesis, and a transcriptomic pattern consistent with epithelial reprograming and promotion of the Paneth cell-like signature in the proximal colon and immune dysregulation characteristic of CD. The observed inflammatory response was associated with persistently increased abundance of Ruminococcus gnavus, Erysipelatoclostridium ramosum, Faecalimonas umbilicate, Blautia hominis, Clostridium butyricum, and C. paraputrificum and unexpected growth of toxigenic C. difficile, which was below the detection level in the community used for inoculation. Our study provides the first evidence that the transfer of a dysbiotic community from CD patients can lead to spontaneous inflammatory changes in the colon of xGF mice and identifies a signature microbial community capable of promoting colonization of pathogenic and conditionally pathogenic bacteria.
Keywords: Crohn’s ileocolitis; fecal microbiome transplant; germ-free mice; inflammation; microbiota.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


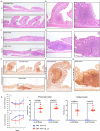

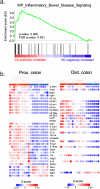

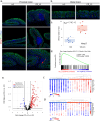
References
-
- de Lange KM, Moutsianas L, Lee JC, de Lange KM, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49(2):256–261. doi:10.1038/ng.3760. - DOI - PMC - PubMed
-
- Kuenzig ME, Fung SG, Marderfeld L, Mak JWY, Kaplan GG, Ng SC, Wilson DC, Cameron F, Henderson P, Kotze PG. et al. Twenty-first century trends in the global epidemiology of pediatric-onset inflammatory bowel disease: systematic review. Gastroenterology. 2022;162(4):1147–1159.e4. doi:10.1053/j.gastro.2021.12.282. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
