The genome sequence of the dark-edged bee fly, Bombylius major (Linnaeus, 1758)
- PMID: 38533437
- PMCID: PMC10964003
- DOI: 10.12688/wellcomeopenres.19804.1
The genome sequence of the dark-edged bee fly, Bombylius major (Linnaeus, 1758)
Abstract
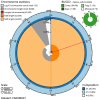
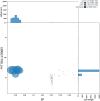
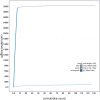
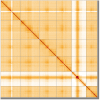
We present a genome assembly from an individual male Bombylius major (the dark-edged bee fly; Arthropoda; Insecta; Diptera; Bombyliidae). The genome sequence is 304.3 megabases in span. The whole assembly is scaffolded into 7 chromosomal pseudomolecules, including the X and Y sex chromosomes. The mitochondrial genome has also been assembled and is 17.8 kilobases in length. Gene annotation of this assembly on Ensembl identified 10,852 protein coding genes.
Keywords: Bombylius major; Diptera; chromosomal; dark-edged bee fly; genome sequence.
Copyright: © 2023 Lawniczak MKN et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
The genome sequence of the dotted bee-fly, Bombylius discolor (Mikan, 1796).Wellcome Open Res. 2022 Dec 21;7:306. doi: 10.12688/wellcomeopenres.18614.1. eCollection 2022. Wellcome Open Res. 2022. PMID: 37124844 Free PMC article.
-
The genome sequence of a conopid fly, Myopa testacea (Linnaeus, 1767).Wellcome Open Res. 2024 Feb 29;9:99. doi: 10.12688/wellcomeopenres.20647.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 38689759 Free PMC article.
-
The genome sequence of the Downland Villa bee-fly, Villa cingulata (Meigen, 1804).Wellcome Open Res. 2023 Nov 14;8:526. doi: 10.12688/wellcomeopenres.20123.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39108663 Free PMC article.
-
The genome sequence of a bluebottle fly, Calliphora vicina (Linnaeus, 1758).Wellcome Open Res. 2024 Jun 24;9:335. doi: 10.12688/wellcomeopenres.22469.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39473873 Free PMC article.
-
The genome sequence of a tachinid fly, Cistogaster globosa (Fabricius, 1775).Wellcome Open Res. 2023 Oct 13;8:462. doi: 10.12688/wellcomeopenres.19924.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39092424 Free PMC article.
Cited by
-
The Ribosomal Operon Database: A Full-Length rDNA Operon Database Derived From Genome Assemblies.Mol Ecol Resour. 2025 Jan;25(1):e14031. doi: 10.1111/1755-0998.14031. Epub 2024 Oct 21. Mol Ecol Resour. 2025. PMID: 39428982 Free PMC article.
-
Do the "big four" orders of insects comprise evolutionarily significant higher taxa with coherent patterns of selection on protein-coding genes?Evol Lett. 2025 Mar 6;9(3):355-366. doi: 10.1093/evlett/qraf005. eCollection 2025 Jun. Evol Lett. 2025. PMID: 40487868 Free PMC article.
References
LinkOut - more resources
Full Text Sources

