Inhibitory protein-protein interactions of the SIRT1 deacetylase are choreographed by post-translational modification
- PMID: 38533551
- PMCID: PMC10966392
- DOI: 10.1002/pro.4938
Inhibitory protein-protein interactions of the SIRT1 deacetylase are choreographed by post-translational modification
Abstract
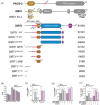
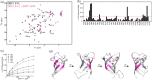
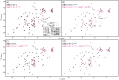
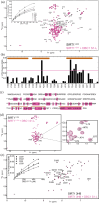
Regulation of SIRT1 activity is vital to energy homeostasis and plays important roles in many diseases. We previously showed that insulin triggers the epigenetic regulator DBC1 to prime SIRT1 for repression by the multifunctional trafficking protein PACS-2. Here, we show that liver DBC1/PACS-2 regulates the diurnal inhibition of SIRT1, which is critically important for insulin-dependent switch in fuel metabolism from fat to glucose oxidation. We present the x-ray structure of the DBC1 S1-like domain that binds SIRT1 and an NMR characterization of how the SIRT1 N-terminal region engages DBC1. This interaction is inhibited by acetylation of K112 of DBC1 and stimulated by the insulin-dependent phosphorylation of human SIRT1 at S162 and S172, catalyzed sequentially by CK2 and GSK3, resulting in the PACS-2-dependent inhibition of nuclear SIRT1 enzymatic activity and translocation of the deacetylase in the cytoplasm. Finally, we discuss how defects in the DBC1/PACS-2-controlled SIRT1 inhibitory pathway are associated with disease, including obesity and non-alcoholic fatty liver disease.
Keywords: CK2; DBC1; GSK3; NAFLD; PACS‐2; SIRT1; acetylation; insulin signaling; liver metabolism; obesity; post‐translational modification; protein–protein interactions.
© 2024 The Protein Society.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

