Modification of Mesenchymal Stem/Stromal Cell-Derived Small Extracellular Vesicles by Calcitonin Gene Related Peptide (CGRP) Antagonist: Potential Implications for Inflammation and Pain Reversal
- PMID: 38534328
- PMCID: PMC10969778
- DOI: 10.3390/cells13060484
Modification of Mesenchymal Stem/Stromal Cell-Derived Small Extracellular Vesicles by Calcitonin Gene Related Peptide (CGRP) Antagonist: Potential Implications for Inflammation and Pain Reversal
Abstract
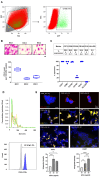
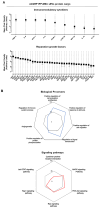
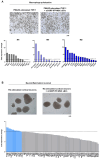
During the progression of knee osteoarthritis (OA), the synovium and infrapatellar fat pad (IFP) can serve as source for Substance P (SP) and calcitonin gene-related peptide (CGRP), two important pain-transmitting, immune, and inflammation modulating neuropeptides. Our previous studies showed that infrapatellar fat pad-derived mesenchymal stem/stromal cells (MSC) acquire a potent immunomodulatory phenotype and actively degrade Substance P via CD10 both in vitro and in vivo. On this basis, our hypothesis is that CD10-bound IFP-MSC sEVs can be engineered to target CGRP while retaining their anti-inflammatory phenotype. Herein, human IFP-MSC cultures were transduced with an adeno-associated virus (AAV) vector carrying a GFP-labelled gene for a CGRP antagonist peptide (aCGRP). The GFP positive aCGRP IFP-MSC were isolated and their sEVs' miRNA and protein cargos were assessed using multiplex methods. Our results showed that purified aCGRP IFP-MSC cultures yielded sEVs with cargo of 147 distinct MSC-related miRNAs. Reactome analysis of miRNAs detected in these sEVs revealed strong involvement in the regulation of target genes involved in pathways that control pain, inflammation and cartilage homeostasis. Protein array of the sEVs cargo demonstrated high presence of key immunomodulatory and reparative proteins. Stimulated macrophages exposed to aCGRP IFP-MSC sEVs demonstrated a switch towards an alternate M2 status. Also, stimulated cortical neurons exposed to aCGRP IFP-MSC sEVs modulate their molecular pain signaling profile. Collectively, our data suggest that yielded sEVs can putatively target CGRP in vivo, while containing potent anti-inflammatory and analgesic cargo, suggesting the promise for novel sEVs-based therapeutic approaches to diseases such as OA.
Keywords: CD10 (neprilysin); analgesia; calcitonin gene-related peptide (CGRP); immunomodulation; infrapatellar fat pad (IFP); mesenchymal stem/stromal cells (MSC); osteoarthritis (OA); small extracellular vesicles (sEVs); synovium.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Saxler G., Löer F., Skumavc M., Pförtner J., Hanesch U. Localization of SP-and CGRP-immunopositive nerve fibers in the hip joint of patients with painful osteoarthritis and of patients with painless failed total hip arthroplasties. Eur. J. Pain. 2007;11:67. doi: 10.1016/j.ejpain.2005.12.011. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

