Exploring Regorafenib Responsiveness and Uncovering Molecular Mechanisms in Recurrent Glioblastoma Tumors through Longitudinal In Vitro Sampling
- PMID: 38534332
- PMCID: PMC10968984
- DOI: 10.3390/cells13060487
Exploring Regorafenib Responsiveness and Uncovering Molecular Mechanisms in Recurrent Glioblastoma Tumors through Longitudinal In Vitro Sampling
Abstract
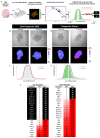
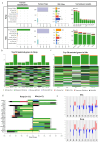
Glioblastoma, a deadly brain tumor, shows limited response to standard therapies like temozolomide (TMZ). Recent findings from the REGOMA trial underscore a significant survival improvement offered by Regorafenib (REGO) in recurrent glioblastoma. Our study aimed to propose a 3D ex vivo drug response precision medicine approach to investigate recurrent glioblastoma sensitivity to REGO and elucidate the underlying molecular mechanisms involved in tumor resistance or responsiveness to treatment. Three-dimensional glioblastoma organoids (GB-EXPs) obtained from 18 patients' resected recurrent glioblastoma tumors were treated with TMZ and REGO. Drug responses were evaluated using NAD(P)H FLIM, stratifying tumors as responders (Resp) or non-responders (NRs). Whole-exome sequencing was performed on 16 tissue samples, and whole-transcriptome analysis on 13 GB-EXPs treated and untreated. We found 35% (n = 9) and 77% (n = 20) of tumors responded to TMZ and REGO, respectively, with no instances of TMZ-Resp being REGO-NRs. Exome analysis revealed a unique mutational profile in REGO-Resp tumors compared to NR tumors. Transcriptome analysis identified distinct expression patterns in Resp and NR tumors, impacting Rho GTPase and NOTCH signaling, known to be involved in drug response. In conclusion, recurrent glioblastoma tumors were more responsive to REGO compared to TMZ treatment. Importantly, our approach enables a comprehensive longitudinal exploration of the molecular changes induced by treatment, unveiling promising biomarkers indicative of drug response.
Keywords: NADP(H) FLIM; Regorafenib; drug response; glioblastoma; organoids.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J.B., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. - DOI - PubMed
-
- Batchelor T.T., Gerstner E.R., Ye X., Desideri S., Duda D.G., Peereboom D., Lesser G.J., Chowdhary S., Wen P.Y., Grossman S., et al. Feasibility, phase I, and phase II studies of tandutinib, an oral platelet-derived growth factor receptor-β tyrosine kinase inhibitor, in patients with recurrent glioblastoma. Neuro Oncol. 2017;19:567–575. doi: 10.1093/neuonc/now185. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

