KIF2A Upregulates PI3K/AKT Signaling through Polo-like Kinase 1 (PLK1) to Affect the Proliferation and Apoptosis Levels of Eriocheir sinensis Spermatogenic Cells
- PMID: 38534420
- PMCID: PMC10968087
- DOI: 10.3390/biology13030149
KIF2A Upregulates PI3K/AKT Signaling through Polo-like Kinase 1 (PLK1) to Affect the Proliferation and Apoptosis Levels of Eriocheir sinensis Spermatogenic Cells
Abstract
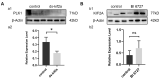
E. sinensis is an animal model for studying the reproduction and development of crustaceans. In this study, we knocked down the Es-Kif2a gene by injecting dsRNA into E. sinensis and inhibited Es-Plk1 gene expression by injecting PLK1 inhibitor BI6727 into E. sinensis. Then, the cell proliferation level, apoptosis level, and PI3K/AKT signaling expression level were detected. Our results showed that the proliferation level of spermatogenic cells decreased, while the apoptosis level increased after Es-Kif2a knockdown or Es-Plk1 inhibition. In order to verify whether these changes are caused by regulating the PI3K/AKT pathway, we detected the expression of PI3K and AKT proteins after Es-Kif2a knockdown or Es-Plk1 inhibition. Western Blot showed that in both the Es-Kif2a knockdown group and the Es-Plk1 inhibition group, the expression of PI3K and AKT proteins decreased. In addition, immunofluorescence showed that Es-KIF2A and Es-PLK1 proteins were co-localized during E. sinensis spermatogenesis. To further explore the upstream and downstream relationship between Es-KIF2A and Es-PLK1, we detected the expression level of Es-PLK1 after Es-Kif2a knockdown as well as the expression level of Es-KIF2A after Es-Plk1 inhibition. Western Blot showed that the expression of Es-PLK1 decreased after Es-Kif2a knockdown, while there was no significant change of Es-KIF2A after Es-Plk1 inhibition, indicating that Es-PLK1 may be a downstream factor of Es-KIF2A. Taken together, these results suggest that Es-KIF2A upregulates the PI3K/AKT signaling pathway through Es-PLK1 during the spermatogenesis of E. sinensis, thereby affecting the proliferation and apoptosis levels of spermatogenic cells.
Keywords: Eriocheir sinensis; KIF2A; PI3K/AKT signaling; PLK1; apoptosis; cell proliferation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








Similar articles
-
Regulation of cell apoptosis and proliferation in pancreatic cancer through PI3K/Akt pathway via Polo-like kinase 1.Oncol Rep. 2016 Jul;36(1):49-56. doi: 10.3892/or.2016.4820. Epub 2016 May 18. Oncol Rep. 2016. PMID: 27220401 Free PMC article.
-
Knockdown of circHIPK3 Facilitates Temozolomide Sensitivity in Glioma by Regulating Cellular Behaviors Through miR-524-5p/KIF2A-Mediated PI3K/AKT Pathway.Cancer Biother Radiopharm. 2021 Sep;36(7):556-567. doi: 10.1089/cbr.2020.3575. Epub 2020 Aug 21. Cancer Biother Radiopharm. 2021. PMID: 32833501
-
Kinesin family member 2A acts as a potential prognostic marker and treatment target via interaction with PI3K/AKT and RhoA/ROCK pathways in acute myeloid leukemia.Oncol Rep. 2022 Jan;47(1):18. doi: 10.3892/or.2021.8229. Epub 2021 Nov 18. Oncol Rep. 2022. PMID: 34792179 Free PMC article.
-
Silencing Kif2a induces apoptosis in squamous cell carcinoma of the oral tongue through inhibition of the PI3K/Akt signaling pathway.Mol Med Rep. 2014 Jan;9(1):273-8. doi: 10.3892/mmr.2013.1804. Epub 2013 Nov 15. Mol Med Rep. 2014. PMID: 24248467
-
Kinesin family member 2A promotes cancer cell viability, mobility, stemness, and chemoresistance to cisplatin by activating the PI3K/AKT/VEGF signaling pathway in non-small cell lung cancer.Am J Transl Res. 2021 Apr 15;13(4):2060-2076. eCollection 2021. Am J Transl Res. 2021. PMID: 34017375 Free PMC article.
Cited by
-
Comprehensive analysis of long non-coding RNA and mRNA expression patterns during seminiferous tubules maturation in Guanzhong dairy goats.BMC Genomics. 2025 Feb 18;26(1):159. doi: 10.1186/s12864-025-11320-7. BMC Genomics. 2025. PMID: 39966702 Free PMC article.
References
-
- Du N.S. Congeneric species of Chinese mitten crab and their English names. Aquat. Sci. Technol. Inf. 1998;25:108–113. doi: 10.16446/j.cnki.1001-1994.1998.03.003. (In Chinese) - DOI
-
- Du N.S., Xue L.Z., Lai W. Histological study on the male reproductive system of Eriocheir sinensis. Acta Zool. Sin. 1988;34:329–333+389–390. (In Chinese)
-
- Du N.S., Xue L.Z., Lai W. Study on Sperm of Eriocheir sinensis II. Spermatogenesis. Oceanol. Limnol. Sin. 1988;19:71–75+103–104. (In Chinese)
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

