Innovations in Antifungal Drug Discovery among Cell Envelope Synthesis Enzymes through Structural Insights
- PMID: 38535180
- PMCID: PMC10970773
- DOI: 10.3390/jof10030171
Innovations in Antifungal Drug Discovery among Cell Envelope Synthesis Enzymes through Structural Insights
Abstract
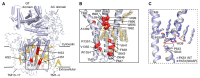
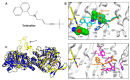
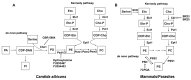
Life-threatening systemic fungal infections occur in immunocompromised patients at an alarming rate. Current antifungal therapies face challenges like drug resistance and patient toxicity, emphasizing the need for new treatments. Membrane-bound enzymes account for a large proportion of current and potential antifungal targets, especially ones that contribute to cell wall and cell membrane biosynthesis. Moreover, structural biology has led to a better understanding of the mechanisms by which these enzymes synthesize their products, as well as the mechanism of action for some antifungals. This review summarizes the structures of several current and potential membrane-bound antifungal targets involved in cell wall and cell membrane biosynthesis and their interactions with known inhibitors or drugs. The proposed mechanisms of action for some molecules, gleaned from detailed inhibitor-protein studeis, are also described, which aids in further rational drug design. Furthermore, some potential membrane-bound antifungal targets with known inhibitors that lack solved structures are discussed, as these might be good enzymes for future structure interrogation.
Keywords: antifungal development; cryo-EM; drug resistance; membrane-bound enzymes; rational drug design; structure biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Natural products targeting the synthesis of β(1,3)-D-glucan and chitin of the fungal cell wall. Existing drugs and recent findings.Phytomedicine. 2021 Jul 15;88:153556. doi: 10.1016/j.phymed.2021.153556. Epub 2021 Mar 27. Phytomedicine. 2021. PMID: 33958276 Review.
-
Recent developments in membrane targeting antifungal agents to mitigate antifungal resistance.RSC Med Chem. 2023 Jun 26;14(9):1603-1628. doi: 10.1039/d3md00151b. eCollection 2023 Sep 19. RSC Med Chem. 2023. PMID: 37731690 Free PMC article. Review.
-
New antifungal agents.Dermatol Clin. 2003 Jul;21(3):565-76. doi: 10.1016/s0733-8635(03)00024-x. Dermatol Clin. 2003. PMID: 12956208 Review.
-
Targeting Unconventional Pathways in Pursuit of Novel Antifungals.Front Mol Biosci. 2021 Jan 12;7:621366. doi: 10.3389/fmolb.2020.621366. eCollection 2020. Front Mol Biosci. 2021. PMID: 33511160 Free PMC article. Review.
-
Antifungal Therapy: New Advances in the Understanding and Treatment of Mycosis.Front Microbiol. 2017 Jan 23;8:36. doi: 10.3389/fmicb.2017.00036. eCollection 2017. Front Microbiol. 2017. PMID: 28167935 Free PMC article. Review.
Cited by
-
In vitro and In silico investigation deciphering novel antifungal activity of endophyte Bacillus velezensis CBMB205 against Fusarium oxysporum.Sci Rep. 2025 Jan 3;15(1):684. doi: 10.1038/s41598-024-77926-1. Sci Rep. 2025. PMID: 39753601 Free PMC article.
-
The small molecule CBR-5884 inhibits the Candida albicans phosphatidylserine synthase.mBio. 2024 May 8;15(5):e0063324. doi: 10.1128/mbio.00633-24. Epub 2024 Apr 9. mBio. 2024. PMID: 38587428 Free PMC article.
-
Garlic-Derived Quorum Sensing Inhibitors: A Novel Strategy Against Fungal Resistance.Drug Des Devel Ther. 2024 Dec 28;18:6413-6426. doi: 10.2147/DDDT.S503302. eCollection 2024. Drug Des Devel Ther. 2024. PMID: 39749188 Free PMC article. Review.
References
-
- Kullberg B., Filler S., Calderone R. Candida and Candidiasis. American Society for Microbiology Press; Washington, DC, USA: 2002.
-
- Morrell M., Fraser V.J., Kollef M.H. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 2005;49:3640–3645. doi: 10.1128/AAC.49.9.3640-3645.2005. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

