Systematic Engineering of Saccharomyces cerevisiae for the De Novo Biosynthesis of Genistein and Glycosylation Derivatives
- PMID: 38535185
- PMCID: PMC10970955
- DOI: 10.3390/jof10030176
Systematic Engineering of Saccharomyces cerevisiae for the De Novo Biosynthesis of Genistein and Glycosylation Derivatives
Abstract
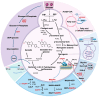
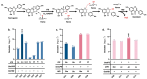
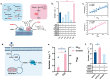
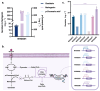
Isoflavones are predominantly found in legumes and play roles in plant defense and prevention of estrogen-related diseases. Genistein is an important isoflavone backbone with various biological activities. In this paper, we describe how a cell factory that can de novo synthesize genistein was constructed in Saccharomyces cerevisiae. Different combinations of isoflavone synthase, cytochrome P450 reductase, and 2-hydroxyisoflavone dehydratase were tested, followed by pathway multicopy integration, to stably de novo synthesize genistein. The catalytic activity of isoflavone synthase was enhanced by heme supply and an increased intracellular NADPH/NADP+ ratio. Redistribution of the malonyl-CoA flow and balance of metabolic fluxes were achieved by adjusting the fatty acid synthesis pathway, yielding 23.33 mg/L genistein. Finally, isoflavone glycosyltransferases were introduced into S. cerevisiae, and the optimized strain produced 15.80 mg/L of genistin or 10.03 mg/L of genistein-8-C-glucoside. This is the first de novo synthesis of genistein-8-C-glucoside in S. cerevisiae, which is advantageous for the green industrial production of isoflavone compounds.
Keywords: genistein; glycosylation; heme; isoflavones; malonyl-CoA.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Engineering Saccharomyces cerevisiae for De Novo Biosynthesis of 3'-Hydroxygenistein.J Agric Food Chem. 2025 Feb 26;73(8):4797-4806. doi: 10.1021/acs.jafc.4c11201. Epub 2025 Feb 13. J Agric Food Chem. 2025. PMID: 39948827
-
Sustainable production of genistin from glycerol by constructing and optimizing Escherichia coli.Metab Eng. 2022 Nov;74:206-219. doi: 10.1016/j.ymben.2022.10.015. Epub 2022 Nov 4. Metab Eng. 2022. PMID: 36336175
-
Modular Engineering of Saccharomyces cerevisiae for De Novo Biosynthesis of Genistein.Microorganisms. 2022 Jul 12;10(7):1402. doi: 10.3390/microorganisms10071402. Microorganisms. 2022. PMID: 35889121 Free PMC article.
-
Recent progress in metabolic engineering of Saccharomyces cerevisiae for the production of malonyl-CoA derivatives.J Biotechnol. 2021 Jan 10;325:83-90. doi: 10.1016/j.jbiotec.2020.11.014. Epub 2020 Dec 2. J Biotechnol. 2021. PMID: 33278463 Review.
-
Genistein.Phytochemistry. 2002 Jun;60(3):205-11. doi: 10.1016/s0031-9422(02)00116-4. Phytochemistry. 2002. PMID: 12031439 Review.
Cited by
-
Advances in synthesizing plant-derived isoflavones and their precursors with multiple pharmacological activities using engineered yeasts.Microb Cell Fact. 2025 Mar 29;24(1):75. doi: 10.1186/s12934-025-02692-2. Microb Cell Fact. 2025. PMID: 40155940 Free PMC article. Review.
References
-
- Jensen S.N., Cady N.M., Shahi S.K., Peterson S.R., Gupta A., Gibson-Corley K.N., Mangalam A.K. Isoflavone diet ameliorates experimental autoimmune encephalomyelitis through modulation of gut bacteria depleted in patients with multiple sclerosis. Sci. Adv. 2021;7:eabd4595. doi: 10.1126/sciadv.abd4595. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

