Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus
- PMID: 38535197
- PMCID: PMC10970960
- DOI: 10.3390/jof10030188
Construction of a Cosmid-Based Ultraefficient Genomic Library System for Filamentous Fungi of the Genus Aspergillus
Abstract
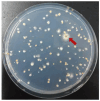
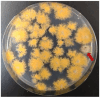
Filamentous fungi of the genus Aspergillus include producers of industrially important organic acids, enzymes, and secondary metabolites, as well as pathogens of many plants and animals. Novel genes in the Aspergillus genome are potentially crucial for the fermentation and drug industries (e.g., agrochemicals and antifungal drugs). A research approach based on classical genetics is effective for identifying functionally unknown genes. During analyses based on classical genetics, mutations must be identified easily and quickly. Herein, we report the development of a cosmid-based plasmid pTOCK1 and the use of a genomic library of Aspergillus nidulans constructed using pTOCK1. The cosmid-based genomic library was used for convenient auxotrophic mutants (pyroA and pabaB), as well as mutants with abnormal colony morphology (gfsA) and yellow conidia (yA), to obtain library clones complementary to these phenotypes. The complementary strain could be obtained through a single transformation, and the cosmid could be rescued. Thus, our cosmid library system can be used to identify the causative gene in a mutant strain.
Keywords: Aspergillus; classical genetics; cosmid vector; genomic library.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Revitalization of a Forward Genetic Screen Identifies Three New Regulators of Fungal Secondary Metabolism in the Genus Aspergillus.mBio. 2017 Sep 5;8(5):e01246-17. doi: 10.1128/mBio.01246-17. mBio. 2017. PMID: 28874473 Free PMC article.
-
Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence.Med Microbiol Immunol. 1998 Oct;187(2):79-89. doi: 10.1007/s004300050077. Med Microbiol Immunol. 1998. PMID: 9832321
-
Bidirectional gene transfer between Aspergillus fumigatus and Aspergillus nidulans.FEMS Microbiol Lett. 1994 Oct 1;122(3):227-31. doi: 10.1111/j.1574-6968.1994.tb07172.x. FEMS Microbiol Lett. 1994. PMID: 7988865
-
Improving extracellular production of food-use enzymes from Aspergillus nidulans.J Biotechnol. 2002 Jun 13;96(1):43-54. doi: 10.1016/s0168-1656(02)00036-6. J Biotechnol. 2002. PMID: 12142142 Review.
-
CRISPR/Cas9-Based Genome Editing and Its Application in Aspergillus Species.J Fungi (Basel). 2022 Apr 30;8(5):467. doi: 10.3390/jof8050467. J Fungi (Basel). 2022. PMID: 35628723 Free PMC article. Review.
References
-
- De Vries R.P., Riley R., Wiebenga A., Aguilar-Osorio G., Amillis S., Uchima C.A., Anderluh G., Asadollahi M., Askin M., Barry K., et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18:28. doi: 10.1186/s13059-017-1151-0. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

