The Peptide LLTRAGL Derived from Rapana venosa Exerts Protective Effect against Inflammatory Bowel Disease in Zebrafish Model by Regulating Multi-Pathways
- PMID: 38535441
- PMCID: PMC10972007
- DOI: 10.3390/md22030100
The Peptide LLTRAGL Derived from Rapana venosa Exerts Protective Effect against Inflammatory Bowel Disease in Zebrafish Model by Regulating Multi-Pathways
Abstract
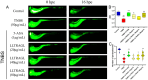
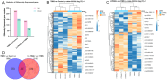
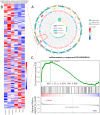
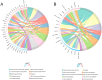
Inflammatory bowel disease (IBD) is a chronic inflammatory bowel disease with unknown pathogenesis which has been gradually considered a public health challenge worldwide. Peptides derived from Rapana venosa have been shown to have an anti-inflammatory effect. In this study, peptide LLTRAGL derived from Rapana venosa was prepared by a solid phase synthesis technique. The protective effects of LLTRAGL were studied in a 2,4,6-trinitrobenzene sulfonic acid (TNBS)-induced zebrafish colitis model. The underlying mechanisms of LLTRAGL were predicted and validated by transcriptome, real-time quantitative PCR assays and molecular docking. The results showed that LLTRAGL reduced the number of macrophages migrating to the intestine, enhanced the frequency and rate of intestinal peristalsis and improved intestinal inflammatory damage. Furthermore, transcriptome analysis indicated the key pathways (NOD-like receptor signal pathway and necroptosis pathway) that link the underlying protective effects of LLTRAGL's molecular mechanisms. In addition, the related genes in these pathways exhibited different expressions after TNBS treatment. Finally, molecular docking techniques further verified the RNA-sequencing results. In summary, LLTRAGL exerted protective effects in the model of TNBS-induced colitis zebrafish. Our findings provide valuable information for the future application of LLTRAGL in IBD.
Keywords: 2,4,6 trinitrobenzene sulfonic acid; LLTRAGL; NOD-like receptor; Rapana venosa; inflammatory bowel disease; necroptosis; transcriptome analysis; zebrafish.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

