Adenine Methylation Enhances the Conformational Flexibility of an RNA Hairpin Tetraloop
- PMID: 38535997
- PMCID: PMC11000223
- DOI: 10.1021/acs.jpcb.4c00522
Adenine Methylation Enhances the Conformational Flexibility of an RNA Hairpin Tetraloop
Abstract
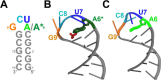
The N6-methyladenosine modification is one of the most abundant post-transcriptional modifications in ribonucleic acid (RNA) molecules. Using molecular dynamics simulations and alchemical free-energy calculations, we studied the structural and energetic implications of incorporating this modification in an adenine mononucleotide and an RNA hairpin structure. At the mononucleotide level, we found that the syn configuration is more favorable than the anti configuration by 2.05 ± 0.15 kcal/mol. The unfavorable effect of methylation was due to the steric overlap between the methyl group and a nitrogen atom in the purine ring. We then probed the effect of methylation in an RNA hairpin structure containing an AUCG tetraloop, which is recognized by a "reader" protein (YTHDC1) to promote transcriptional silencing of long noncoding RNAs. While methylation had no significant conformational effect on the hairpin stem, the methylated tetraloop showed enhanced conformational flexibility compared to the unmethylated tetraloop. The increased flexibility was associated with the outward flipping of two bases (A6 and U7) which formed stacking interactions with each other and with the C8 and G9 bases in the tetraloop, leading to a conformation similar to that in the RNA/reader protein complex. Therefore, methylation-induced conformational flexibility likely facilitates RNA recognition by the reader protein.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Structural effects of m6A modification of the Xist A-repeat AUCG tetraloop and its recognition by YTHDC1.Nucleic Acids Res. 2022 Feb 28;50(4):2350-2362. doi: 10.1093/nar/gkac080. Nucleic Acids Res. 2022. PMID: 35166835 Free PMC article.
-
Dynamics and stability of GCAA tetraloops with 2-aminopurine and purine substitutions.J Biomol Struct Dyn. 2005 Feb;22(4):425-39. doi: 10.1080/07391102.2005.10507014. J Biomol Struct Dyn. 2005. PMID: 15588106
-
Effects of base substitutions in an RNA hairpin from molecular dynamics and free energy simulations.Biophys J. 2003 Dec;85(6):3445-59. doi: 10.1016/S0006-3495(03)74766-3. Biophys J. 2003. PMID: 14645041 Free PMC article.
-
Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):49-67. doi: 10.1002/wrna.1196. Epub 2013 Oct 3. Wiley Interdiscip Rev RNA. 2014. PMID: 24124096 Free PMC article. Review.
-
An RNA folding motif: GNRA tetraloop-receptor interactions.Q Rev Biophys. 2013 Aug;46(3):223-64. doi: 10.1017/S0033583513000048. Epub 2013 Aug 5. Q Rev Biophys. 2013. PMID: 23915736 Review.
Cited by
-
Rationalizing the effects of RNA modifications on protein interactions.Mol Ther Nucleic Acids. 2024 Nov 15;35(4):102391. doi: 10.1016/j.omtn.2024.102391. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39717617 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

