Discovery and characterization of noncanonical E2-conjugating enzymes
- PMID: 38536929
- PMCID: PMC10971424
- DOI: 10.1126/sciadv.adh0123
Discovery and characterization of noncanonical E2-conjugating enzymes
Abstract
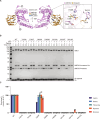
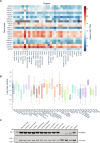
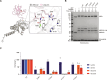
E2-conjugating enzymes (E2s) play a central role in the enzymatic cascade that leads to the attachment of ubiquitin to a substrate. This process, termed ubiquitylation, is required to maintain cellular homeostasis and affects almost all cellular process. By interacting with multiple E3 ligases, E2s dictate the ubiquitylation landscape within the cell. Since its discovery, ubiquitylation has been regarded as a posttranslational modification that specifically targets lysine side chains (canonical ubiquitylation). We used Matrix-Assisted Laser Desorption/Ionization-Time Of Flight Mass Spectrometry to identify and characterize a family of E2s that are instead able to conjugate ubiquitin to serine and/or threonine. We used structural modeling and prediction tools to identify the key activity determinants that these E2s use to interact with ubiquitin as well as their substrates. Our results unveil the missing E2s necessary for noncanonical ubiquitylation, underscoring the adaptability and versatility of ubiquitin modifications.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

