CRIF1 deficiency induces FOXP3LOW inflammatory non-suppressive regulatory T cells, thereby promoting antitumor immunity
- PMID: 38536932
- PMCID: PMC10971410
- DOI: 10.1126/sciadv.adj9600
CRIF1 deficiency induces FOXP3LOW inflammatory non-suppressive regulatory T cells, thereby promoting antitumor immunity
Abstract
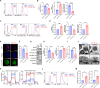
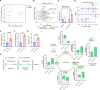
Recently identified human FOXP3lowCD45RA- inflammatory non-suppressive (INS) cells produce proinflammatory cytokines, exhibit reduced suppressiveness, and promote antitumor immunity unlike conventional regulatory T cells (Tregs). In spite of their implication in tumors, the mechanism for generation of FOXP3lowCD45RA- INS cells in vivo is unclear. We showed that the FOXP3lowCD45RA- cells in human tumors demonstrate attenuated expression of CRIF1, a vital mitochondrial regulator. Mice with CRIF1 deficiency in Tregs bore Foxp3lowINS-Tregs with mitochondrial dysfunction and metabolic reprograming. The enhanced glutaminolysis activated α-ketoglutarate-mTORC1 axis, which promoted proinflammatory cytokine expression by inducing EOMES and SATB1 expression. Moreover, chromatin openness of the regulatory regions of the Ifng and Il4 genes was increased, which facilitated EOMES/SATB1 binding. The increased α-ketoglutarate-derived 2-hydroxyglutarate down-regulated Foxp3 expression by methylating the Foxp3 gene regulatory regions. Furthermore, CRIF1 deficiency-induced Foxp3lowINS-Tregs suppressed tumor growth in an IFN-γ-dependent manner. Thus, CRIF1 deficiency-mediated mitochondrial dysfunction results in the induction of Foxp3lowINS-Tregs including FOXP3lowCD45RA- cells that promote antitumor immunity.
Figures





References
-
- Sakaguchi S., Mikami N., Wing J. B., Tanaka A., Ichiyama K., Ohkura N., Regulatory T cells and human disease. Annu. Rev. Immunol. 38, 541–566 (2020). - PubMed
-
- O’Callaghan D. S., Rexhepaj E., Gately K., Coate L., Delaney D., O’Donnell D. M., Kay E., O’Connell F., Gallagher W. M., O’Byrne K. J., Tumour islet Foxp3+ T-cell infiltration predicts poor outcome in nonsmall cell lung cancer. Eur. Respir. J. 46, 1762–1772 (2015). - PubMed
-
- Saleh R., Elkord E., FoxP3+ T regulatory cells in cancer: Prognostic biomarkers and therapeutic targets. Cancer Lett. 490, 174–185 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

