Information Theoretic Study of COVID-19 Genome
- PMID: 38539735
- PMCID: PMC10968974
- DOI: 10.3390/e26030223
Information Theoretic Study of COVID-19 Genome
Abstract
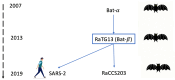
In this paper, we analyse the genome sequence of COVID-19 on a information point of view, and we compare that with past and present genomes. We use the powerful tool of joint complexity in order to quantify the similarities measured between the various potential parent genomes. The tool has a computing complexity of several orders of magnitude below the classic Smith-Waterman algorithm and would allow it to be used on a larger scale.
Keywords: COVID-19; genome; joint complexity; pattern matching.
Conflict of interest statement
The author declares no conflict of interest.
Figures








References
-
- Jacquet P., Milioris D., Szpankowski W. Classification of Markov sources through joint string complexity: Theory and experiments; Proceedings of the 2013 IEEE International Symposium on Information Theory; Istanbul, Turkey. 7–12 July 2013; pp. 2289–2293.
-
- Milioris D. Topic Detection and Classification in Social Networks. Springer; Berlin/Heidelberg, Germany: 2018. Joint Sequence Complexity: Introduction and Theory; pp. 21–56.
-
- Burnside G., Milioris D., Jacquet P. One Day in Twitter: Topic Detection Via Joint Complexity; Proceedings of the SNOW 2014 Data Challenge; Seoul, Republic of Korea. 8 April 2014.
LinkOut - more resources
Full Text Sources
Miscellaneous

