An Integrated Approach Including CRISPR/Cas9-Mediated Nanopore Sequencing, Mate Pair Sequencing, and Cytogenomic Methods to Characterize Complex Structural Rearrangements in Acute Myeloid Leukemia
- PMID: 38540211
- PMCID: PMC10968562
- DOI: 10.3390/biomedicines12030598
An Integrated Approach Including CRISPR/Cas9-Mediated Nanopore Sequencing, Mate Pair Sequencing, and Cytogenomic Methods to Characterize Complex Structural Rearrangements in Acute Myeloid Leukemia
Abstract
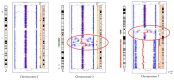
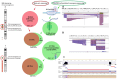
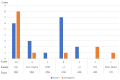
Complex structural chromosome abnormalities such as chromoanagenesis have been reported in acute myeloid leukemia (AML). They are usually not well characterized by conventional genetic methods, and the characterization of chromoanagenesis structural abnormalities from short-read sequencing still presents challenges. Here, we characterized complex structural abnormalities involving chromosomes 2, 3, and 7 in an AML patient using an integrated approach including CRISPR/Cas9-mediated nanopore sequencing, mate pair sequencing (MPseq), and SNP microarray analysis along with cytogenetic methods. SNP microarray analysis revealed chromoanagenesis involving chromosomes 3 and 7, and a pseudotricentric chromosome 7 was revealed by cytogenetic methods. MPseq revealed 138 structural variants (SVs) as putative junctions of complex rearrangements involving chromosomes 2, 3, and 7, which led to 16 novel gene fusions and 33 truncated genes. Thirty CRISPR RNA (crRNA) sequences were designed to map 29 SVs, of which 27 (93.1%) were on-target based on CRISPR/Cas9 crRNA nanopore sequencing. In addition to simple SVs, complex SVs involving over two breakpoints were also revealed. Twenty-one SVs (77.8% of the on-target SVs) were also revealed by MPseq with shared SV breakpoints. Approximately three-quarters of breakpoints were located within genes, especially intronic regions, and one-quarter of breakpoints were intergenic. Alu and LINE repeat elements were frequent among breakpoints. Amplification of the chromosome 7 centromere was also detected by nanopore sequencing. Given the high amplification of the chromosome 7 centromere, extra chromosome 7 centromere sequences (tricentric), and more gains than losses of genomic material, chromoanasynthesis and chromothripsis may be responsible for forming this highly complex structural abnormality. We showed this combination approach's value in characterizing complex structural abnormalities for clinical and research applications. Characterization of these complex structural chromosome abnormalities not only will help understand the molecular mechanisms responsible for the process of chromoanagenesis, but also may identify specific molecular targets and their impact on therapy and overall survival.
Keywords: CRISPR/Cas9; acute myeloid leukemia; chromoanagenesis; complex structural abnormalities; mate pair sequencing; nanopore sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

