Subepithelial Stromal Cells: Their Roles and Interactions with Intestinal Epithelial Cells during Gut Mucosal Homeostasis and Regeneration
- PMID: 38540281
- PMCID: PMC10968202
- DOI: 10.3390/biomedicines12030668
Subepithelial Stromal Cells: Their Roles and Interactions with Intestinal Epithelial Cells during Gut Mucosal Homeostasis and Regeneration
Abstract
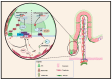
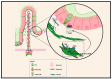
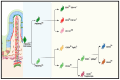
Intestinal epithelial cell activities during homeostasis and regeneration are well described, but their potential interactions with stromal cells remain unresolved. Exploring the functions of these heterogeneous intestinal mesenchymal stromal cells (iMSCs) remains challenging. This difficulty is due to the lack of specific markers for most functionally homogenous subpopulations. In recent years, however, novel clustering techniques such as single-cell RNA sequencing (scRNA-seq), fluorescence-activated cell sorting (FACS), confocal microscope, and computational remodeling of intestinal anatomy have helped identify and characterize some specific iMSC subsets. These methods help researchers learn more about the localization and functions of iMSC populations during intestinal morphogenic and homeostatic conditions. Consequently, it is imperative to understand the cellular pathways that regulate their activation and how they interact with surrounding cellular components, particularly during intestinal epithelial regeneration after mucosal injury. This review provides insights into the spatial distribution and functions of identified iMSC subtypes. It focuses on their involvement in intestinal morphogenesis, homeostasis, and regeneration. We reviewed related signaling mechanisms implicated during epithelial and subepithelial stromal cell crosstalk. Future research should focus on elucidating the molecular intermediates of these regulatory pathways to open a new frontier for potential therapeutic targets that can alleviate intestinal mucosa-related injuries.
Keywords: epithelial–mesenchymal interactions; intestinal stem-cell niche; mesenchymal stromal cells; subepithelial gradient factors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

