Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors
- PMID: 38540380
- PMCID: PMC10969806
- DOI: 10.3390/genes15030321
Understanding the Variability of 22q11.2 Deletion Syndrome: The Role of Epigenetic Factors
Abstract
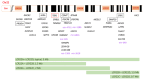
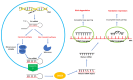
Initially described as a triad of immunodeficiency, congenital heart defects and hypoparathyroidism, 22q11.2 deletion syndrome (22q11.2DS) now encompasses a great amount of abnormalities involving different systems. Approximately 85% of patients share a 3 Mb 22q11.2 region of hemizygous deletion in which 46 protein-coding genes are included. However, the hemizygosity of the genes of this region cannot fully explain the clinical phenotype and the phenotypic variability observed among patients. Additional mutations in genes located outside the deleted region, leading to "dual diagnosis", have been described in 1% of patients. In some cases, the hemizygosity of the 22q11.2 region unmasks autosomal recessive conditions due to additional mutations on the non-deleted allele. Some of the deleted genes play a crucial role in gene expression regulation pathways, involving the whole genome. Typical miRNA expression patterns have been identified in 22q11.2DS, due to an alteration in miRNA biogenesis, affecting the expression of several target genes. Also, a methylation epi-signature in CpG islands differentiating patients from controls has been defined. Herein, we summarize the evidence on the genetic and epigenetic mechanisms implicated in the pathogenesis of the clinical manifestations of 22q11.2 DS. The review of the literature confirms the hypothesis that the 22q11.2DS phenotype results from a network of interactions between deleted protein-coding genes and altered epigenetic regulation.
Keywords: 22q11.2 deletion syndrome; CpG islands; epigenetics; methylation; micro-RNAs.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Grati F.R., Molina Gomes D., Ferreira J.C., Dupont C., Alesi V., Gouas L., Horelli-Kuitunen N., Choy K.W., García-Herrero S., de la Vega A.G., et al. Prevalence of recurrent pathogenic microdeletions and microduplications in over 9500 pregnancies. Prenat. Diagn. 2015;35:801–809. doi: 10.1002/pd.4613. - DOI - PubMed
-
- Gross S.J., Stosic M., McDonald-McGinn D.M., Bassett A.S., Norvez A., Dhamankar R., Kobara K., Kirkizlar E., Zimmermann B., Wayham N., et al. Clinical experience with single-nucleotide polymorphism-based non-invasive prenatal screening for 22q11.2 deletion syndrome. Ultrasound Obstet. Gynecol. 2016;47:177–183. doi: 10.1002/uog.15754. - DOI - PMC - PubMed
-
- Blagojevic C., Heung T., Theriault M., Tomita-Mitchell A., Chakraborty P., Kernohan K., Bulman D.E., Bassett A.S. Estimate of the contemporary live-birth prevalence of recurrent 22q11.2 deletions: A cross-sectional analysis from population-based newborn screening. CMAJ Open. 2021;9:E802–E809. doi: 10.9778/cmajo.20200294. - DOI - PMC - PubMed
-
- Bevilacqua E., Jani J.C., Chaoui R., Suk E.A., Palma-Dias R., Ko T.M., Warsof S., Stokowski R., Jones K.J., Grati F.R., et al. Performance of a targeted cell-free DNA prenatal test for 22q11.2 deletion in a large clinical cohort. Ultrasound Obstet. Gynecol. 2021;58:597–602. doi: 10.1002/uog.23699. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

