Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data
- PMID: 38540410
- PMCID: PMC10970060
- DOI: 10.3390/genes15030351
Population Structure and Selection Signal Analysis of Nanyang Cattle Based on Whole-Genome Sequencing Data
Abstract
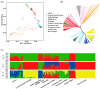
With a rich breeding history, Nanyang cattle (NY cattle) have undergone extensive natural and artificial selection, resulting in distinctive traits such as high fertility, excellent meat quality, and disease resistance. This makes them an ideal model for studying the mechanisms of environmental adaptability. To assess the population structure and genetic diversity of NY cattle, we performed whole-genome resequencing on 30 individuals. These data were then compared with published whole-genome resequencing data from 432 cattle globally. The results indicate that the genetic structure of NY cattle is significantly different from European commercial breeds and is more similar to North-Central Chinese breeds. Furthermore, among all breeds, NY cattle exhibit the highest genetic diversity and the lowest population inbreeding levels. A genome-wide selection signal analysis of NY cattle and European commercial breeds using Fst, θπ-ratio, and θπ methods revealed significant selection signals in genes associated with reproductive performance and immunity. Our functional annotation analysis suggests that these genes may be responsible for reproduction (MAP2K2, PGR, and GSE1), immune response (NCOA2, HSF1, and PAX5), and olfaction (TAS1R3). We provide a comprehensive overview of sequence variations in the NY cattle genome, revealing insights into the population structure and genetic diversity of NY cattle. Additionally, we identify candidate genes associated with important economic traits, offering valuable references for future conservation and breeding efforts of NY cattle.
Keywords: Nanyang cattle; genetic diversity; population structure; whole-genome resequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Analysis of Population Structure and Selective Signatures for Milk Production Traits in Xinjiang Brown Cattle and Chinese Simmental Cattle.Int J Mol Sci. 2025 Feb 25;26(5):2003. doi: 10.3390/ijms26052003. Int J Mol Sci. 2025. PMID: 40076627 Free PMC article.
-
Whole-genome sequencing reveals genomic diversity and selection signatures in Xia'nan cattle.BMC Genomics. 2024 Jun 5;25(1):559. doi: 10.1186/s12864-024-10463-3. BMC Genomics. 2024. PMID: 38840048 Free PMC article.
-
Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data.BMC Genomics. 2021 Jan 9;22(1):43. doi: 10.1186/s12864-020-07340-0. BMC Genomics. 2021. PMID: 33421990 Free PMC article. Review.
-
Insights into Adaption and Growth Evolution: Genome-Wide Copy Number Variation Analysis in Chinese Hainan Yellow Cattle Using Whole-Genome Re-Sequencing Data.Int J Mol Sci. 2024 Nov 6;25(22):11919. doi: 10.3390/ijms252211919. Int J Mol Sci. 2024. PMID: 39595990 Free PMC article.
-
Beef Cattle Genome Project: Advances in Genome Sequencing, Assembly, and Functional Genes Discovery.Int J Mol Sci. 2024 Jun 28;25(13):7147. doi: 10.3390/ijms25137147. Int J Mol Sci. 2024. PMID: 39000250 Free PMC article. Review.
Cited by
-
Analysis of Population Structure and Selective Signatures for Milk Production Traits in Xinjiang Brown Cattle and Chinese Simmental Cattle.Int J Mol Sci. 2025 Feb 25;26(5):2003. doi: 10.3390/ijms26052003. Int J Mol Sci. 2025. PMID: 40076627 Free PMC article.
-
Examination of homozygosity runs and selection signatures in native goat breeds of Henan, China.BMC Genomics. 2024 Dec 7;25(1):1184. doi: 10.1186/s12864-024-11098-0. BMC Genomics. 2024. PMID: 39643897 Free PMC article.
-
Genome-Wide Analysis of Copy Number Variations in Three Populations of Nanyang Cattle Using Whole-Genome Resequencing.Genes (Basel). 2025 May 12;16(5):568. doi: 10.3390/genes16050568. Genes (Basel). 2025. PMID: 40428390 Free PMC article.
-
Analysis of genomic selection characteristics of local cattle breeds in Gansu.BMC Genomics. 2025 Jul 1;26(1):574. doi: 10.1186/s12864-025-11753-0. BMC Genomics. 2025. PMID: 40597588 Free PMC article.
-
Genetic and Evolutionary Analysis of Ake Chicken: New Insights into China's Sole Indigenous Naked-Neck Chicken Breed.Int J Mol Sci. 2025 May 6;26(9):4399. doi: 10.3390/ijms26094399. Int J Mol Sci. 2025. PMID: 40362637 Free PMC article.
References
-
- Chinese Academy of Agricultural Sciences. 2020. [(accessed on 5 November 2023)]. Available online: https://zypc.nahs.org.cn/pzml/index.html.
Publication types
MeSH terms
Grants and funding
- 32202636/the National Natural Science Foundation of China
- 221100110200/the Major Scientific and Technological Special Project of Henan Province
- 32002168/the National Natural Science Foundation of China
- 2018YFD0501704/Application and Demonstration of High Quality Beef Cattle High Efficient and Safe Feeding Technology
- CARS-36/Third National Census of Livestock and Poultry Genetic Resources
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

