Genome-Wide DNA Methylation Analysis and Functional Validation of Litter Size Traits in Jining Grey Goats
- PMID: 38540412
- PMCID: PMC10970512
- DOI: 10.3390/genes15030353
Genome-Wide DNA Methylation Analysis and Functional Validation of Litter Size Traits in Jining Grey Goats
Abstract
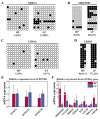
DNA methylation (DNAm) is associated with the reproductive system. However, the genetic mechanism through which DNAm regulates gene expression and thus affects litter size in goats is unclear. Therefore, in the present work, genome-wide DNAm profiles of HP and LP Jining Grey goat ovary tissues were comprehensively analyzed via WGBS, and RNA-Seq data were combined to identify candidate genes associated with litter size traits in the Jining Grey goat. Finally, BSP and RT-qPCR were used to verify the sequencing results of the key genes. Notably, the DNMT genes were downregulated at the expression level in the HP group. Both groups exhibited comparable levels of methylation. A total of 976 differentially methylated regions (DMRs) (973 DMRs for CG and 3 DMRs for CHG) and 310 differentially methylated genes (DMGs) were identified in this study. Through integration of WGBS and RNA-Seq data, we identified 59 differentially methylated and differentially expressed genes (DEGs) and ultimately screened 8 key DMGs (9 DMRS) associated with litter size traits in Jining Grey goats (SERPINB2: chr24_62258801_62259000, NDRG4: chr18_27599201_27599400, CFAP43: chr26_27046601_27046800, LRP1B. chr2_79720201_79720400, EPHA6: chr1_40088601_40088800, TTC29: chr17_59385801_59386000, PDE11A: chr2_117418601_117418800 and PGF: chr10_ 16913801_16914000 and chr10_16916401_16916600). In summary, our research comprehensively analyzed the genome-wide DNAm profiles of HP and LP Jining Grey goat ovary tissues. The data findings suggest that DNAm in goat ovaries may play an important role in determining litter size.
Keywords: DNA methylation; gene expression; goat ovary; prolificacy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Potential Candidate Genes Associated with Litter Size in Goats: A Review.Animals (Basel). 2025 Jan 2;15(1):82. doi: 10.3390/ani15010082. Animals (Basel). 2025. PMID: 39795025 Free PMC article. Review.
-
Genome-wide analysis of DNA Methylation profiles on sheep ovaries associated with prolificacy using whole-genome Bisulfite sequencing.BMC Genomics. 2017 Oct 2;18(1):759. doi: 10.1186/s12864-017-4068-9. BMC Genomics. 2017. PMID: 28969601 Free PMC article.
-
Genome-wide profile in DNA methylation in goat ovaries of two different litter size populations.J Anim Physiol Anim Nutr (Berl). 2022 Mar;106(2):239-249. doi: 10.1111/jpn.13600. Epub 2021 Jul 1. J Anim Physiol Anim Nutr (Berl). 2022. PMID: 34212445
-
Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity.Gene. 2016 May 10;582(1):69-76. doi: 10.1016/j.gene.2016.01.047. Epub 2016 Feb 4. Gene. 2016. PMID: 26851539
-
Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers.Genes (Basel). 2023 May 29;14(6):1183. doi: 10.3390/genes14061183. Genes (Basel). 2023. PMID: 37372363 Free PMC article. Review.
Cited by
-
Potential Candidate Genes Associated with Litter Size in Goats: A Review.Animals (Basel). 2025 Jan 2;15(1):82. doi: 10.3390/ani15010082. Animals (Basel). 2025. PMID: 39795025 Free PMC article. Review.
-
Genomic advancements in goat breeding: enhancing productivity, disease resistance, and sustainability in India's rural economy.Mamm Genome. 2025 Sep;36(3):761-786. doi: 10.1007/s00335-025-10138-8. Epub 2025 May 28. Mamm Genome. 2025. PMID: 40434651 Review.
-
Genomic and Molecular Mechanisms of Goat Environmental Adaptation.Biology (Basel). 2025 Jun 5;14(6):654. doi: 10.3390/biology14060654. Biology (Basel). 2025. PMID: 40563905 Free PMC article. Review.
References
-
- Wang R., Wang Z., Wang X., Li Y., Qu L., Lan X. A novel 4-bp insertion within the goat CFAP43 gene and its association with litter size. Small Rumin. Res. 2021;202:106456. doi: 10.1016/j.smallrumres.2021.106456. - DOI
Publication types
MeSH terms
Grants and funding
- CXGC2023F10/Agricultural Scientific and Technological Innovation Project of Shandong Academy of Agricultural Sciences
- 32372851/Molecular mechanism of hair follicle development in high-quality fine-wool sheep revealed by single-cell transcriptome-based study
- YDZX2023012/Creation and Application of Healthy and Efficient Feed for Yak and Tibetan Sheep in Haibei Prefecture
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

